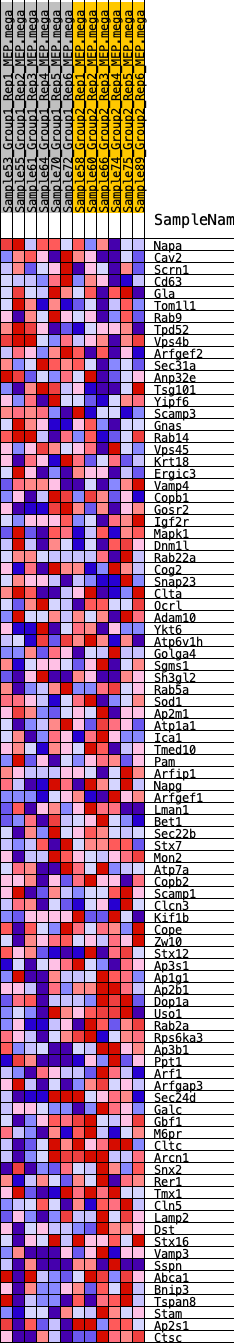
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
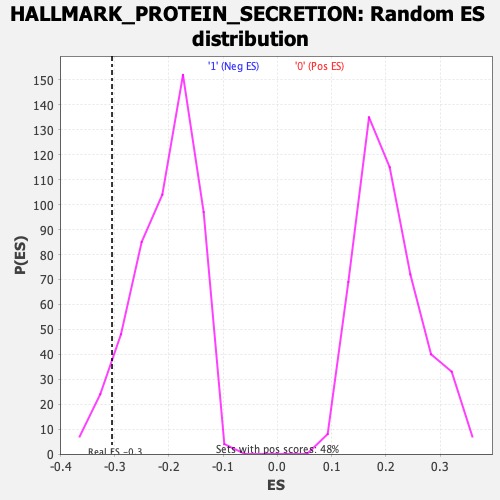
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.30496767 |
| Normalized Enrichment Score (NES) | -1.4703002 |
| Nominal p-value | 0.07293666 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.487 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Napa | 1150 | 0.439 | -0.0299 | No |
| 2 | Cav2 | 1308 | 0.419 | -0.0095 | No |
| 3 | Scrn1 | 1600 | 0.386 | 0.0017 | No |
| 4 | Cd63 | 1816 | 0.367 | 0.0156 | No |
| 5 | Gla | 3679 | 0.236 | -0.0652 | No |
| 6 | Tom1l1 | 3827 | 0.229 | -0.0572 | No |
| 7 | Rab9 | 4290 | 0.204 | -0.0673 | No |
| 8 | Tpd52 | 4359 | 0.201 | -0.0571 | No |
| 9 | Vps4b | 4532 | 0.193 | -0.0529 | No |
| 10 | Arfgef2 | 4574 | 0.191 | -0.0420 | No |
| 11 | Sec31a | 5204 | 0.161 | -0.0638 | No |
| 12 | Anp32e | 5297 | 0.156 | -0.0579 | No |
| 13 | Tsg101 | 5489 | 0.148 | -0.0577 | No |
| 14 | Yipf6 | 5498 | 0.148 | -0.0480 | No |
| 15 | Scamp3 | 6065 | 0.125 | -0.0690 | No |
| 16 | Gnas | 6323 | 0.116 | -0.0744 | No |
| 17 | Rab14 | 6722 | 0.100 | -0.0883 | No |
| 18 | Vps45 | 7045 | 0.088 | -0.0991 | No |
| 19 | Krt18 | 7790 | 0.061 | -0.1336 | No |
| 20 | Ergic3 | 8021 | 0.053 | -0.1419 | No |
| 21 | Vamp4 | 8031 | 0.053 | -0.1388 | No |
| 22 | Copb1 | 8097 | 0.050 | -0.1388 | No |
| 23 | Gosr2 | 8955 | 0.022 | -0.1818 | No |
| 24 | Igf2r | 9088 | 0.018 | -0.1875 | No |
| 25 | Mapk1 | 9257 | 0.012 | -0.1954 | No |
| 26 | Dnm1l | 9452 | 0.006 | -0.2051 | No |
| 27 | Rab22a | 9686 | -0.001 | -0.2171 | No |
| 28 | Cog2 | 9830 | -0.006 | -0.2242 | No |
| 29 | Snap23 | 9942 | -0.010 | -0.2293 | No |
| 30 | Clta | 9985 | -0.011 | -0.2307 | No |
| 31 | Ocrl | 9995 | -0.012 | -0.2304 | No |
| 32 | Adam10 | 10045 | -0.013 | -0.2321 | No |
| 33 | Ykt6 | 10049 | -0.013 | -0.2313 | No |
| 34 | Atp6v1h | 10150 | -0.017 | -0.2354 | No |
| 35 | Golga4 | 10167 | -0.017 | -0.2350 | No |
| 36 | Sgms1 | 10233 | -0.020 | -0.2371 | No |
| 37 | Sh3gl2 | 10296 | -0.022 | -0.2388 | No |
| 38 | Rab5a | 10318 | -0.023 | -0.2383 | No |
| 39 | Sod1 | 10747 | -0.036 | -0.2581 | No |
| 40 | Ap2m1 | 11292 | -0.055 | -0.2827 | No |
| 41 | Atp1a1 | 11293 | -0.055 | -0.2789 | No |
| 42 | Ica1 | 11335 | -0.056 | -0.2772 | No |
| 43 | Tmed10 | 11386 | -0.058 | -0.2758 | No |
| 44 | Pam | 11583 | -0.066 | -0.2815 | No |
| 45 | Arfip1 | 11602 | -0.067 | -0.2779 | No |
| 46 | Napg | 11756 | -0.071 | -0.2810 | No |
| 47 | Arfgef1 | 12218 | -0.090 | -0.2988 | Yes |
| 48 | Lman1 | 12296 | -0.092 | -0.2965 | Yes |
| 49 | Bet1 | 12299 | -0.092 | -0.2904 | Yes |
| 50 | Sec22b | 12327 | -0.093 | -0.2854 | Yes |
| 51 | Stx7 | 12402 | -0.096 | -0.2827 | Yes |
| 52 | Mon2 | 12493 | -0.099 | -0.2806 | Yes |
| 53 | Atp7a | 12626 | -0.105 | -0.2803 | Yes |
| 54 | Copb2 | 12629 | -0.105 | -0.2732 | Yes |
| 55 | Scamp1 | 12707 | -0.109 | -0.2698 | Yes |
| 56 | Clcn3 | 12785 | -0.111 | -0.2662 | Yes |
| 57 | Kif1b | 12981 | -0.118 | -0.2683 | Yes |
| 58 | Cope | 13189 | -0.125 | -0.2705 | Yes |
| 59 | Zw10 | 13240 | -0.127 | -0.2645 | Yes |
| 60 | Stx12 | 13381 | -0.133 | -0.2627 | Yes |
| 61 | Ap3s1 | 13548 | -0.139 | -0.2619 | Yes |
| 62 | Ap1g1 | 13744 | -0.147 | -0.2620 | Yes |
| 63 | Ap2b1 | 13827 | -0.151 | -0.2559 | Yes |
| 64 | Dop1a | 13850 | -0.152 | -0.2467 | Yes |
| 65 | Uso1 | 13883 | -0.154 | -0.2379 | Yes |
| 66 | Rab2a | 13945 | -0.157 | -0.2303 | Yes |
| 67 | Rps6ka3 | 14101 | -0.162 | -0.2273 | Yes |
| 68 | Ap3b1 | 14565 | -0.184 | -0.2388 | Yes |
| 69 | Ppt1 | 14693 | -0.189 | -0.2326 | Yes |
| 70 | Arf1 | 14878 | -0.198 | -0.2287 | Yes |
| 71 | Arfgap3 | 14883 | -0.198 | -0.2154 | Yes |
| 72 | Sec24d | 14896 | -0.199 | -0.2024 | Yes |
| 73 | Galc | 14978 | -0.203 | -0.1928 | Yes |
| 74 | Gbf1 | 15129 | -0.211 | -0.1862 | Yes |
| 75 | M6pr | 15303 | -0.220 | -0.1802 | Yes |
| 76 | Cltc | 15308 | -0.220 | -0.1654 | Yes |
| 77 | Arcn1 | 15440 | -0.227 | -0.1568 | Yes |
| 78 | Snx2 | 15785 | -0.244 | -0.1580 | Yes |
| 79 | Rer1 | 15891 | -0.251 | -0.1463 | Yes |
| 80 | Tmx1 | 15956 | -0.254 | -0.1323 | Yes |
| 81 | Cln5 | 16272 | -0.275 | -0.1300 | Yes |
| 82 | Lamp2 | 16408 | -0.284 | -0.1177 | Yes |
| 83 | Dst | 16764 | -0.307 | -0.1152 | Yes |
| 84 | Stx16 | 16795 | -0.310 | -0.0956 | Yes |
| 85 | Vamp3 | 17328 | -0.352 | -0.0992 | Yes |
| 86 | Sspn | 17425 | -0.361 | -0.0796 | Yes |
| 87 | Abca1 | 17531 | -0.369 | -0.0599 | Yes |
| 88 | Bnip3 | 17652 | -0.381 | -0.0401 | Yes |
| 89 | Tspan8 | 17893 | -0.405 | -0.0249 | Yes |
| 90 | Stam | 18283 | -0.450 | -0.0144 | Yes |
| 91 | Ap2s1 | 18284 | -0.451 | 0.0164 | Yes |
| 92 | Ctsc | 18752 | -0.539 | 0.0289 | Yes |