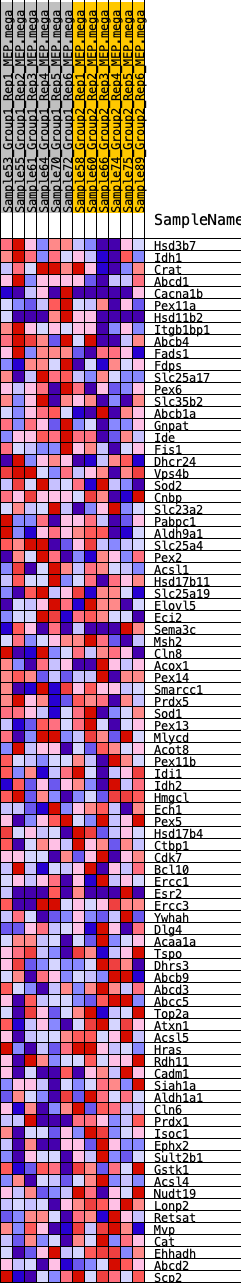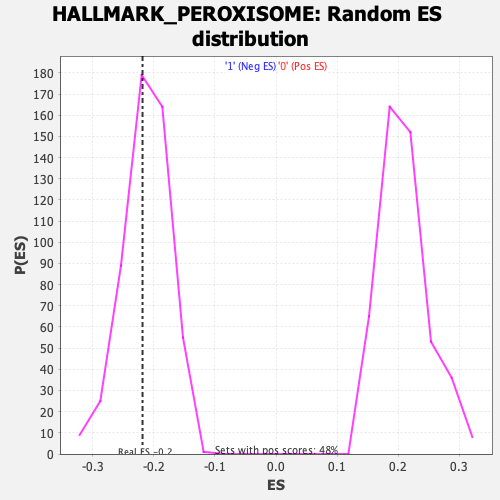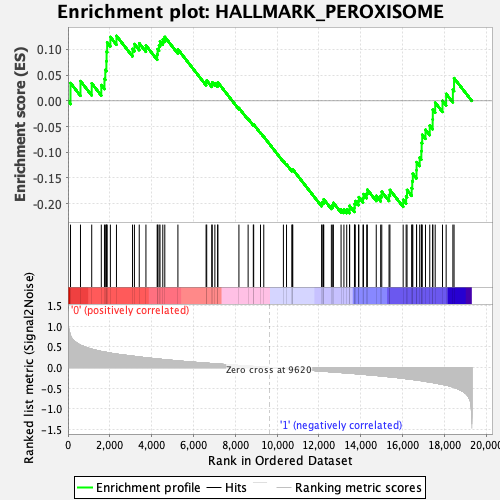
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.21835048 |
| Normalized Enrichment Score (NES) | -1.0251898 |
| Nominal p-value | 0.3908046 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd3b7 | 120 | 0.771 | 0.0347 | No |
| 2 | Idh1 | 597 | 0.538 | 0.0384 | No |
| 3 | Crat | 1134 | 0.443 | 0.0340 | No |
| 4 | Abcd1 | 1592 | 0.386 | 0.0307 | No |
| 5 | Cacna1b | 1743 | 0.374 | 0.0428 | No |
| 6 | Pex11a | 1783 | 0.370 | 0.0604 | No |
| 7 | Hsd11b2 | 1835 | 0.365 | 0.0771 | No |
| 8 | Itgb1bp1 | 1845 | 0.364 | 0.0959 | No |
| 9 | Abcb4 | 1872 | 0.362 | 0.1138 | No |
| 10 | Fads1 | 2026 | 0.347 | 0.1242 | No |
| 11 | Fdps | 2317 | 0.325 | 0.1264 | No |
| 12 | Slc25a17 | 3085 | 0.273 | 0.1010 | No |
| 13 | Pex6 | 3176 | 0.268 | 0.1105 | No |
| 14 | Slc35b2 | 3406 | 0.253 | 0.1120 | No |
| 15 | Abcb1a | 3726 | 0.233 | 0.1078 | No |
| 16 | Gnpat | 4264 | 0.205 | 0.0907 | No |
| 17 | Ide | 4284 | 0.205 | 0.1006 | No |
| 18 | Fis1 | 4352 | 0.201 | 0.1078 | No |
| 19 | Dhcr24 | 4399 | 0.199 | 0.1160 | No |
| 20 | Vps4b | 4532 | 0.193 | 0.1193 | No |
| 21 | Sod2 | 4628 | 0.188 | 0.1244 | No |
| 22 | Cnbp | 5255 | 0.158 | 0.1002 | No |
| 23 | Slc23a2 | 6602 | 0.105 | 0.0357 | No |
| 24 | Pabpc1 | 6632 | 0.104 | 0.0397 | No |
| 25 | Aldh9a1 | 6874 | 0.095 | 0.0322 | No |
| 26 | Slc25a4 | 6895 | 0.094 | 0.0361 | No |
| 27 | Pex2 | 7022 | 0.089 | 0.0343 | No |
| 28 | Acsl1 | 7154 | 0.084 | 0.0319 | No |
| 29 | Hsd17b11 | 7165 | 0.084 | 0.0359 | No |
| 30 | Slc25a19 | 8172 | 0.048 | -0.0139 | No |
| 31 | Elovl5 | 8612 | 0.034 | -0.0349 | No |
| 32 | Eci2 | 8860 | 0.026 | -0.0464 | No |
| 33 | Sema3c | 8878 | 0.025 | -0.0460 | No |
| 34 | Msh2 | 9204 | 0.014 | -0.0622 | No |
| 35 | Cln8 | 9360 | 0.009 | -0.0698 | No |
| 36 | Acox1 | 10299 | -0.022 | -0.1174 | No |
| 37 | Pex14 | 10448 | -0.027 | -0.1237 | No |
| 38 | Smarcc1 | 10702 | -0.035 | -0.1350 | No |
| 39 | Prdx5 | 10729 | -0.036 | -0.1344 | No |
| 40 | Sod1 | 10747 | -0.036 | -0.1334 | No |
| 41 | Pex13 | 12126 | -0.087 | -0.2005 | No |
| 42 | Mlycd | 12153 | -0.088 | -0.1972 | No |
| 43 | Acot8 | 12216 | -0.090 | -0.1956 | No |
| 44 | Pex11b | 12228 | -0.090 | -0.1914 | No |
| 45 | Idi1 | 12592 | -0.104 | -0.2048 | No |
| 46 | Idh2 | 12657 | -0.107 | -0.2024 | No |
| 47 | Hmgcl | 12688 | -0.108 | -0.1983 | No |
| 48 | Ech1 | 13056 | -0.120 | -0.2110 | No |
| 49 | Pex5 | 13192 | -0.125 | -0.2114 | Yes |
| 50 | Hsd17b4 | 13326 | -0.131 | -0.2114 | Yes |
| 51 | Ctbp1 | 13461 | -0.136 | -0.2111 | Yes |
| 52 | Cdk7 | 13466 | -0.136 | -0.2041 | Yes |
| 53 | Bcl10 | 13696 | -0.145 | -0.2084 | Yes |
| 54 | Ercc1 | 13700 | -0.145 | -0.2008 | Yes |
| 55 | Esr2 | 13735 | -0.147 | -0.1948 | Yes |
| 56 | Ercc3 | 13891 | -0.154 | -0.1947 | Yes |
| 57 | Ywhah | 13906 | -0.155 | -0.1872 | Yes |
| 58 | Dlg4 | 14105 | -0.163 | -0.1889 | Yes |
| 59 | Acaa1a | 14127 | -0.164 | -0.1813 | Yes |
| 60 | Tspo | 14283 | -0.171 | -0.1803 | Yes |
| 61 | Dhrs3 | 14312 | -0.172 | -0.1726 | Yes |
| 62 | Abcb9 | 14738 | -0.191 | -0.1846 | Yes |
| 63 | Abcd3 | 14950 | -0.202 | -0.1849 | Yes |
| 64 | Abcc5 | 15002 | -0.204 | -0.1767 | Yes |
| 65 | Top2a | 15350 | -0.222 | -0.1830 | Yes |
| 66 | Atxn1 | 15392 | -0.225 | -0.1732 | Yes |
| 67 | Acsl5 | 16025 | -0.259 | -0.1923 | Yes |
| 68 | Hras | 16167 | -0.269 | -0.1854 | Yes |
| 69 | Rdh11 | 16210 | -0.272 | -0.1732 | Yes |
| 70 | Cadm1 | 16431 | -0.285 | -0.1695 | Yes |
| 71 | Siah1a | 16460 | -0.287 | -0.1558 | Yes |
| 72 | Aldh1a1 | 16486 | -0.289 | -0.1418 | Yes |
| 73 | Cln6 | 16662 | -0.301 | -0.1349 | Yes |
| 74 | Prdx1 | 16668 | -0.302 | -0.1191 | Yes |
| 75 | Isoc1 | 16815 | -0.310 | -0.1103 | Yes |
| 76 | Ephx2 | 16894 | -0.317 | -0.0976 | Yes |
| 77 | Sult2b1 | 16909 | -0.317 | -0.0814 | Yes |
| 78 | Gstk1 | 16936 | -0.321 | -0.0658 | Yes |
| 79 | Acsl4 | 17092 | -0.334 | -0.0561 | Yes |
| 80 | Nudt19 | 17296 | -0.349 | -0.0482 | Yes |
| 81 | Lonp2 | 17435 | -0.362 | -0.0361 | Yes |
| 82 | Retsat | 17442 | -0.363 | -0.0172 | Yes |
| 83 | Mvp | 17556 | -0.372 | -0.0034 | Yes |
| 84 | Cat | 17906 | -0.407 | 0.0001 | Yes |
| 85 | Ehhadh | 18080 | -0.425 | 0.0136 | Yes |
| 86 | Abcd2 | 18402 | -0.468 | 0.0217 | Yes |
| 87 | Scp2 | 18458 | -0.477 | 0.0442 | Yes |