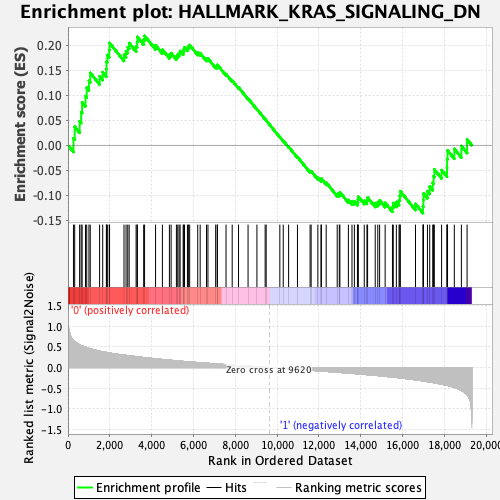
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.21837631 |
| Normalized Enrichment Score (NES) | 0.87491614 |
| Nominal p-value | 0.68604654 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Copz2 | 261 | 0.657 | 0.0138 | Yes |
| 2 | Gamt | 320 | 0.628 | 0.0370 | Yes |
| 3 | Idua | 556 | 0.552 | 0.0478 | Yes |
| 4 | Mast3 | 628 | 0.532 | 0.0663 | Yes |
| 5 | Ryr1 | 675 | 0.523 | 0.0858 | Yes |
| 6 | Pdk2 | 836 | 0.490 | 0.0979 | Yes |
| 7 | Tex15 | 895 | 0.480 | 0.1149 | Yes |
| 8 | Kcnd1 | 1002 | 0.462 | 0.1287 | Yes |
| 9 | Htr1d | 1063 | 0.454 | 0.1445 | Yes |
| 10 | Thrb | 1511 | 0.396 | 0.1378 | Yes |
| 11 | Coq8a | 1659 | 0.381 | 0.1460 | Yes |
| 12 | Hsd11b2 | 1835 | 0.365 | 0.1521 | Yes |
| 13 | Kmt2d | 1840 | 0.365 | 0.1671 | Yes |
| 14 | Zbtb16 | 1881 | 0.361 | 0.1801 | Yes |
| 15 | Synpo | 1970 | 0.353 | 0.1903 | Yes |
| 16 | Mx2 | 1987 | 0.351 | 0.2041 | Yes |
| 17 | Epha5 | 2678 | 0.299 | 0.1807 | Yes |
| 18 | Ybx2 | 2776 | 0.293 | 0.1879 | Yes |
| 19 | Grid2 | 2857 | 0.288 | 0.1957 | Yes |
| 20 | Sphk2 | 2924 | 0.283 | 0.2041 | Yes |
| 21 | Ccdc106 | 3256 | 0.262 | 0.1978 | Yes |
| 22 | Vps50 | 3305 | 0.259 | 0.2061 | Yes |
| 23 | Clstn3 | 3317 | 0.258 | 0.2163 | Yes |
| 24 | Tnni3 | 3618 | 0.240 | 0.2107 | Yes |
| 25 | Htr1b | 3661 | 0.237 | 0.2184 | Yes |
| 26 | Slc25a23 | 4186 | 0.209 | 0.1998 | No |
| 27 | Cpa2 | 4514 | 0.193 | 0.1909 | No |
| 28 | Slc16a7 | 4848 | 0.177 | 0.1809 | No |
| 29 | Ryr2 | 4930 | 0.174 | 0.1840 | No |
| 30 | Slc29a3 | 5183 | 0.162 | 0.1776 | No |
| 31 | Rsad2 | 5247 | 0.159 | 0.1810 | No |
| 32 | Sptbn2 | 5311 | 0.156 | 0.1842 | No |
| 33 | Ptprj | 5360 | 0.154 | 0.1881 | No |
| 34 | Tcf7l1 | 5506 | 0.147 | 0.1867 | No |
| 35 | Kcnn1 | 5533 | 0.146 | 0.1914 | No |
| 36 | Efhd1 | 5571 | 0.145 | 0.1956 | No |
| 37 | Gpr19 | 5711 | 0.139 | 0.1941 | No |
| 38 | Gp1ba | 5756 | 0.138 | 0.1976 | No |
| 39 | Atp4a | 5817 | 0.135 | 0.2001 | No |
| 40 | Lgals7 | 6203 | 0.120 | 0.1850 | No |
| 41 | Myo15a | 6319 | 0.116 | 0.1839 | No |
| 42 | Dtnb | 6623 | 0.104 | 0.1724 | No |
| 43 | Macroh2a2 | 6691 | 0.101 | 0.1732 | No |
| 44 | Slc12a3 | 7060 | 0.088 | 0.1577 | No |
| 45 | Edar | 7135 | 0.085 | 0.1574 | No |
| 46 | Brdt | 7143 | 0.085 | 0.1605 | No |
| 47 | Nr4a2 | 7556 | 0.070 | 0.1420 | No |
| 48 | Tenm2 | 7850 | 0.059 | 0.1292 | No |
| 49 | Asb7 | 8153 | 0.049 | 0.1155 | No |
| 50 | Serpinb2 | 8609 | 0.035 | 0.0933 | No |
| 51 | Celsr2 | 9033 | 0.019 | 0.0721 | No |
| 52 | Slc6a3 | 9426 | 0.007 | 0.0520 | No |
| 53 | Fggy | 9475 | 0.005 | 0.0497 | No |
| 54 | Cd80 | 10128 | -0.016 | 0.0164 | No |
| 55 | Prkn | 10291 | -0.022 | 0.0089 | No |
| 56 | Mthfr | 10548 | -0.030 | -0.0032 | No |
| 57 | Mfsd6 | 10971 | -0.044 | -0.0233 | No |
| 58 | Zc2hc1c | 11575 | -0.066 | -0.0520 | No |
| 59 | Tgm1 | 11625 | -0.068 | -0.0517 | No |
| 60 | P2rx6 | 11946 | -0.080 | -0.0651 | No |
| 61 | Ypel1 | 12095 | -0.086 | -0.0692 | No |
| 62 | Plag1 | 12117 | -0.086 | -0.0667 | No |
| 63 | Tfcp2l1 | 12344 | -0.094 | -0.0745 | No |
| 64 | Arpp21 | 12871 | -0.114 | -0.0972 | No |
| 65 | Bard1 | 12971 | -0.118 | -0.0974 | No |
| 66 | Tent5c | 12999 | -0.119 | -0.0938 | No |
| 67 | Klk8 | 13406 | -0.134 | -0.1094 | No |
| 68 | Cdkal1 | 13578 | -0.140 | -0.1125 | No |
| 69 | Selenop | 13692 | -0.145 | -0.1123 | No |
| 70 | Capn9 | 13839 | -0.152 | -0.1136 | No |
| 71 | Msh5 | 13862 | -0.152 | -0.1084 | No |
| 72 | Entpd7 | 13877 | -0.154 | -0.1027 | No |
| 73 | Nrip2 | 14165 | -0.166 | -0.1107 | No |
| 74 | Gtf3c5 | 14293 | -0.171 | -0.1102 | No |
| 75 | Egf | 14324 | -0.173 | -0.1045 | No |
| 76 | Nos1 | 14689 | -0.188 | -0.1156 | No |
| 77 | Tg | 14812 | -0.194 | -0.1139 | No |
| 78 | Ngb | 14899 | -0.199 | -0.1100 | No |
| 79 | Fgfr3 | 15163 | -0.213 | -0.1148 | No |
| 80 | Sgk1 | 15513 | -0.230 | -0.1234 | No |
| 81 | Btg2 | 15551 | -0.232 | -0.1156 | No |
| 82 | Col2a1 | 15699 | -0.241 | -0.1132 | No |
| 83 | Cyp39a1 | 15828 | -0.246 | -0.1096 | No |
| 84 | Bmpr1b | 15864 | -0.248 | -0.1011 | No |
| 85 | Stag3 | 15888 | -0.250 | -0.0918 | No |
| 86 | Prodh | 16614 | -0.298 | -0.1171 | No |
| 87 | Dcc | 16971 | -0.323 | -0.1222 | No |
| 88 | Tgfb2 | 16990 | -0.325 | -0.1096 | No |
| 89 | Lfng | 16997 | -0.325 | -0.0963 | No |
| 90 | Snn | 17186 | -0.342 | -0.0918 | No |
| 91 | Pde6b | 17294 | -0.349 | -0.0828 | No |
| 92 | Nr6a1 | 17438 | -0.362 | -0.0751 | No |
| 93 | Cpeb3 | 17478 | -0.366 | -0.0619 | No |
| 94 | Thnsl2 | 17514 | -0.368 | -0.0484 | No |
| 95 | Sidt1 | 17862 | -0.402 | -0.0496 | No |
| 96 | Abcg4 | 18120 | -0.429 | -0.0451 | No |
| 97 | Zfp112 | 18126 | -0.430 | -0.0274 | No |
| 98 | Mefv | 18146 | -0.432 | -0.0104 | No |
| 99 | Itgb1bp2 | 18470 | -0.478 | -0.0072 | No |
| 100 | Skil | 18808 | -0.556 | -0.0016 | No |
| 101 | Camk1d | 19081 | -0.658 | 0.0118 | No |

