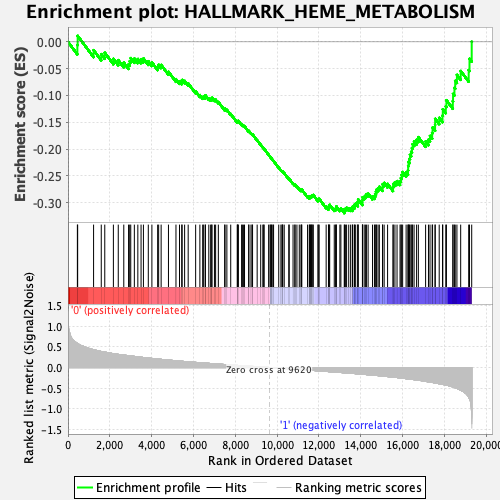
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEME_METABOLISM |
| Enrichment Score (ES) | -0.31943154 |
| Normalized Enrichment Score (NES) | -1.3337603 |
| Nominal p-value | 0.15564202 |
| FDR q-value | 0.97339606 |
| FWER p-Value | 0.759 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mboat2 | 448 | 0.581 | -0.0058 | No |
| 2 | Sidt2 | 462 | 0.579 | 0.0112 | No |
| 3 | C3 | 1223 | 0.429 | -0.0155 | No |
| 4 | Rnf123 | 1590 | 0.386 | -0.0229 | No |
| 5 | Uros | 1758 | 0.373 | -0.0203 | No |
| 6 | Igsf3 | 2170 | 0.335 | -0.0316 | No |
| 7 | Tspan5 | 2403 | 0.317 | -0.0341 | No |
| 8 | Gde1 | 2667 | 0.300 | -0.0387 | No |
| 9 | Slc6a8 | 2895 | 0.285 | -0.0419 | No |
| 10 | Ppox | 2950 | 0.281 | -0.0362 | No |
| 11 | Fbxo9 | 3000 | 0.278 | -0.0303 | No |
| 12 | Mpp1 | 3172 | 0.268 | -0.0311 | No |
| 13 | Mgst3 | 3339 | 0.257 | -0.0319 | No |
| 14 | Slc66a2 | 3493 | 0.248 | -0.0324 | No |
| 15 | Gmps | 3606 | 0.241 | -0.0310 | No |
| 16 | Snca | 3839 | 0.228 | -0.0362 | No |
| 17 | Trim58 | 4011 | 0.220 | -0.0384 | No |
| 18 | Icam4 | 4286 | 0.205 | -0.0465 | No |
| 19 | Tmcc2 | 4334 | 0.202 | -0.0428 | No |
| 20 | Tns1 | 4453 | 0.197 | -0.0430 | No |
| 21 | Atp6v0a1 | 4803 | 0.179 | -0.0558 | No |
| 22 | Pigq | 5162 | 0.162 | -0.0696 | No |
| 23 | Marchf2 | 5331 | 0.155 | -0.0737 | No |
| 24 | Ranbp10 | 5438 | 0.150 | -0.0747 | No |
| 25 | Ncoa4 | 5456 | 0.150 | -0.0710 | No |
| 26 | Bsg | 5576 | 0.145 | -0.0728 | No |
| 27 | Slc7a11 | 5746 | 0.138 | -0.0775 | No |
| 28 | Tmem9b | 6103 | 0.123 | -0.0923 | No |
| 29 | Mocos | 6305 | 0.117 | -0.0993 | No |
| 30 | Agpat4 | 6439 | 0.111 | -0.1029 | No |
| 31 | Pcx | 6486 | 0.110 | -0.1019 | No |
| 32 | Bach1 | 6563 | 0.106 | -0.1027 | No |
| 33 | Tal1 | 6568 | 0.106 | -0.0997 | No |
| 34 | Trak2 | 6732 | 0.100 | -0.1052 | No |
| 35 | Smox | 6820 | 0.096 | -0.1068 | No |
| 36 | Ctns | 6872 | 0.095 | -0.1066 | No |
| 37 | Optn | 6882 | 0.094 | -0.1042 | No |
| 38 | Acp5 | 7002 | 0.090 | -0.1077 | No |
| 39 | Htra2 | 7058 | 0.088 | -0.1079 | No |
| 40 | Slc11a2 | 7188 | 0.083 | -0.1121 | No |
| 41 | Xpo7 | 7489 | 0.072 | -0.1256 | No |
| 42 | Rnf19a | 7509 | 0.071 | -0.1244 | No |
| 43 | Nr3c1 | 7596 | 0.068 | -0.1269 | No |
| 44 | Gclc | 7780 | 0.062 | -0.1345 | No |
| 45 | Alad | 8094 | 0.050 | -0.1494 | No |
| 46 | Gapvd1 | 8100 | 0.050 | -0.1481 | No |
| 47 | Fbxo7 | 8119 | 0.049 | -0.1475 | No |
| 48 | Ell2 | 8146 | 0.049 | -0.1474 | No |
| 49 | Gata1 | 8300 | 0.044 | -0.1541 | No |
| 50 | E2f2 | 8338 | 0.043 | -0.1547 | No |
| 51 | Abcb6 | 8396 | 0.041 | -0.1564 | No |
| 52 | Klf3 | 8440 | 0.040 | -0.1575 | No |
| 53 | Vezf1 | 8642 | 0.034 | -0.1670 | No |
| 54 | Osbp2 | 8646 | 0.034 | -0.1661 | No |
| 55 | Marchf8 | 8749 | 0.030 | -0.1705 | No |
| 56 | Rbm38 | 8804 | 0.028 | -0.1725 | No |
| 57 | Eif2ak1 | 8824 | 0.027 | -0.1727 | No |
| 58 | Foxj2 | 9044 | 0.019 | -0.1835 | No |
| 59 | Riok3 | 9213 | 0.014 | -0.1919 | No |
| 60 | Slc22a4 | 9315 | 0.010 | -0.1969 | No |
| 61 | P4ha2 | 9355 | 0.009 | -0.1987 | No |
| 62 | Rhag | 9378 | 0.008 | -0.1996 | No |
| 63 | Rcl1 | 9592 | 0.001 | -0.2106 | No |
| 64 | Blvrb | 9664 | -0.000 | -0.2144 | No |
| 65 | Slc25a38 | 9672 | -0.000 | -0.2147 | No |
| 66 | Bnip3l | 9728 | -0.002 | -0.2175 | No |
| 67 | Aldh1l1 | 9749 | -0.003 | -0.2185 | No |
| 68 | Add1 | 9800 | -0.005 | -0.2209 | No |
| 69 | Dcun1d1 | 9829 | -0.006 | -0.2222 | No |
| 70 | Sec14l1 | 10076 | -0.014 | -0.2347 | No |
| 71 | Selenbp1 | 10180 | -0.018 | -0.2395 | No |
| 72 | Hebp1 | 10231 | -0.020 | -0.2415 | No |
| 73 | Cir1 | 10255 | -0.021 | -0.2421 | No |
| 74 | Nfe2l1 | 10269 | -0.021 | -0.2421 | No |
| 75 | Narf | 10338 | -0.023 | -0.2450 | No |
| 76 | Mark3 | 10557 | -0.030 | -0.2555 | No |
| 77 | Endod1 | 10576 | -0.031 | -0.2555 | No |
| 78 | Epor | 10760 | -0.037 | -0.2639 | No |
| 79 | Top1 | 10843 | -0.040 | -0.2670 | No |
| 80 | Adipor1 | 10859 | -0.040 | -0.2665 | No |
| 81 | Lrp10 | 10945 | -0.043 | -0.2697 | No |
| 82 | Daam1 | 11058 | -0.047 | -0.2741 | No |
| 83 | Hdgf | 11137 | -0.050 | -0.2767 | No |
| 84 | Foxo3 | 11167 | -0.051 | -0.2766 | No |
| 85 | Gypc | 11177 | -0.051 | -0.2756 | No |
| 86 | Rbm5 | 11453 | -0.061 | -0.2881 | No |
| 87 | Tnrc6b | 11539 | -0.064 | -0.2906 | No |
| 88 | Usp15 | 11540 | -0.064 | -0.2886 | No |
| 89 | Lmo2 | 11590 | -0.066 | -0.2891 | No |
| 90 | Bmp2k | 11608 | -0.067 | -0.2880 | No |
| 91 | Ypel5 | 11648 | -0.068 | -0.2880 | No |
| 92 | Sptb | 11660 | -0.069 | -0.2865 | No |
| 93 | Tfdp2 | 11705 | -0.070 | -0.2866 | No |
| 94 | Synj1 | 11725 | -0.070 | -0.2855 | No |
| 95 | Fbxo34 | 11955 | -0.080 | -0.2950 | No |
| 96 | Nnt | 11962 | -0.080 | -0.2929 | No |
| 97 | Car1 | 12008 | -0.082 | -0.2928 | No |
| 98 | Kat2b | 12343 | -0.094 | -0.3074 | No |
| 99 | Mfhas1 | 12454 | -0.098 | -0.3101 | No |
| 100 | Map2k3 | 12468 | -0.098 | -0.3078 | No |
| 101 | Car2 | 12485 | -0.099 | -0.3057 | No |
| 102 | Ctsb | 12512 | -0.100 | -0.3040 | No |
| 103 | Ppp2r5b | 12730 | -0.110 | -0.3120 | No |
| 104 | Clcn3 | 12785 | -0.111 | -0.3114 | No |
| 105 | Mxi1 | 12822 | -0.112 | -0.3099 | No |
| 106 | Xk | 12830 | -0.113 | -0.3069 | No |
| 107 | Tent5c | 12999 | -0.119 | -0.3120 | No |
| 108 | Khnyn | 13047 | -0.119 | -0.3109 | No |
| 109 | Add2 | 13212 | -0.126 | -0.3156 | Yes |
| 110 | Pdzk1ip1 | 13215 | -0.126 | -0.3119 | Yes |
| 111 | Picalm | 13273 | -0.128 | -0.3110 | Yes |
| 112 | Epb41 | 13320 | -0.130 | -0.3094 | Yes |
| 113 | Nek7 | 13422 | -0.134 | -0.3106 | Yes |
| 114 | Mospd1 | 13513 | -0.137 | -0.3111 | Yes |
| 115 | Ucp2 | 13596 | -0.141 | -0.3111 | Yes |
| 116 | Lpin2 | 13606 | -0.142 | -0.3073 | Yes |
| 117 | Btrc | 13686 | -0.144 | -0.3070 | Yes |
| 118 | Rap1gap | 13701 | -0.145 | -0.3034 | Yes |
| 119 | Aldh6a1 | 13754 | -0.148 | -0.3016 | Yes |
| 120 | Dcaf10 | 13858 | -0.152 | -0.3023 | Yes |
| 121 | Slc2a1 | 13859 | -0.152 | -0.2977 | Yes |
| 122 | Arl2bp | 13873 | -0.153 | -0.2937 | Yes |
| 123 | Abcg2 | 14078 | -0.162 | -0.2995 | Yes |
| 124 | Slc30a1 | 14079 | -0.162 | -0.2946 | Yes |
| 125 | Nfe2 | 14083 | -0.162 | -0.2898 | Yes |
| 126 | Pgls | 14160 | -0.165 | -0.2888 | Yes |
| 127 | Dcaf11 | 14218 | -0.168 | -0.2866 | Yes |
| 128 | Nudt4 | 14268 | -0.170 | -0.2840 | Yes |
| 129 | Kdm7a | 14349 | -0.174 | -0.2829 | Yes |
| 130 | Fech | 14560 | -0.184 | -0.2883 | Yes |
| 131 | Blvra | 14651 | -0.187 | -0.2873 | Yes |
| 132 | Acsl6 | 14688 | -0.188 | -0.2835 | Yes |
| 133 | H1f0 | 14725 | -0.190 | -0.2796 | Yes |
| 134 | Rad23a | 14756 | -0.192 | -0.2753 | Yes |
| 135 | Mkrn1 | 14825 | -0.195 | -0.2729 | Yes |
| 136 | Hmbs | 14886 | -0.198 | -0.2701 | Yes |
| 137 | Glrx5 | 15039 | -0.206 | -0.2717 | Yes |
| 138 | Gclm | 15042 | -0.206 | -0.2656 | Yes |
| 139 | Sdcbp | 15116 | -0.210 | -0.2630 | Yes |
| 140 | Arhgef12 | 15280 | -0.218 | -0.2649 | Yes |
| 141 | Myl4 | 15538 | -0.231 | -0.2713 | Yes |
| 142 | Btg2 | 15551 | -0.232 | -0.2649 | Yes |
| 143 | Cast | 15631 | -0.237 | -0.2618 | Yes |
| 144 | Slc6a9 | 15732 | -0.242 | -0.2597 | Yes |
| 145 | Psmd9 | 15875 | -0.249 | -0.2595 | Yes |
| 146 | Tcea1 | 15918 | -0.252 | -0.2540 | Yes |
| 147 | Htatip2 | 15954 | -0.254 | -0.2482 | Yes |
| 148 | Minpp1 | 16000 | -0.258 | -0.2427 | Yes |
| 149 | Hagh | 16166 | -0.268 | -0.2431 | Yes |
| 150 | Cdc27 | 16255 | -0.274 | -0.2394 | Yes |
| 151 | Cpox | 16265 | -0.274 | -0.2316 | Yes |
| 152 | Epb42 | 16278 | -0.275 | -0.2238 | Yes |
| 153 | Ackr1 | 16331 | -0.279 | -0.2181 | Yes |
| 154 | Bpgm | 16351 | -0.280 | -0.2106 | Yes |
| 155 | Lamp2 | 16408 | -0.284 | -0.2049 | Yes |
| 156 | Tspo2 | 16446 | -0.286 | -0.1981 | Yes |
| 157 | Klf1 | 16476 | -0.288 | -0.1909 | Yes |
| 158 | Dmtn | 16547 | -0.292 | -0.1856 | Yes |
| 159 | Asns | 16667 | -0.302 | -0.1827 | Yes |
| 160 | Tfrc | 16751 | -0.306 | -0.1777 | Yes |
| 161 | Ccdc28a | 17098 | -0.334 | -0.1856 | Yes |
| 162 | Ank1 | 17235 | -0.346 | -0.1822 | Yes |
| 163 | Spta1 | 17308 | -0.350 | -0.1754 | Yes |
| 164 | Ezh1 | 17411 | -0.361 | -0.1697 | Yes |
| 165 | Prdx2 | 17431 | -0.362 | -0.1597 | Yes |
| 166 | Urod | 17551 | -0.371 | -0.1546 | Yes |
| 167 | Ermap | 17555 | -0.372 | -0.1435 | Yes |
| 168 | Slc10a3 | 17748 | -0.391 | -0.1416 | Yes |
| 169 | Cat | 17906 | -0.407 | -0.1375 | Yes |
| 170 | Ctse | 17920 | -0.408 | -0.1257 | Yes |
| 171 | Kel | 18063 | -0.424 | -0.1203 | Yes |
| 172 | Cdr2 | 18093 | -0.427 | -0.1088 | Yes |
| 173 | Slc4a1 | 18399 | -0.467 | -0.1106 | Yes |
| 174 | Tyr | 18411 | -0.469 | -0.0969 | Yes |
| 175 | Fn3k | 18476 | -0.480 | -0.0856 | Yes |
| 176 | Ubac1 | 18513 | -0.489 | -0.0727 | Yes |
| 177 | Ccnd3 | 18595 | -0.505 | -0.0615 | Yes |
| 178 | Trim10 | 18775 | -0.545 | -0.0543 | Yes |
| 179 | Atg4a | 19158 | -0.713 | -0.0526 | Yes |
| 180 | Alas2 | 19197 | -0.757 | -0.0316 | Yes |
| 181 | Rhd | 19302 | -1.226 | 0.0003 | Yes |