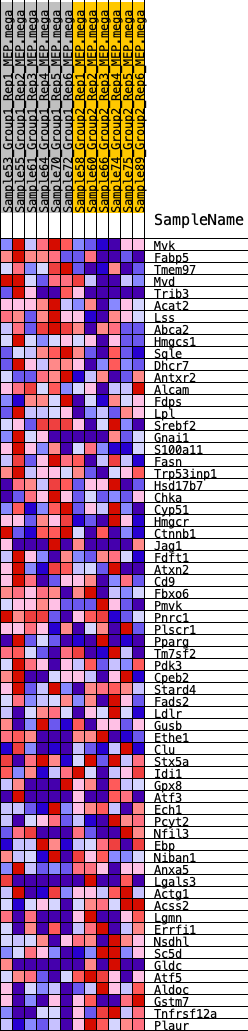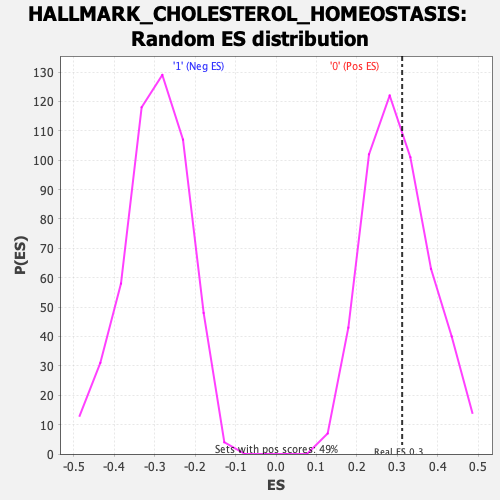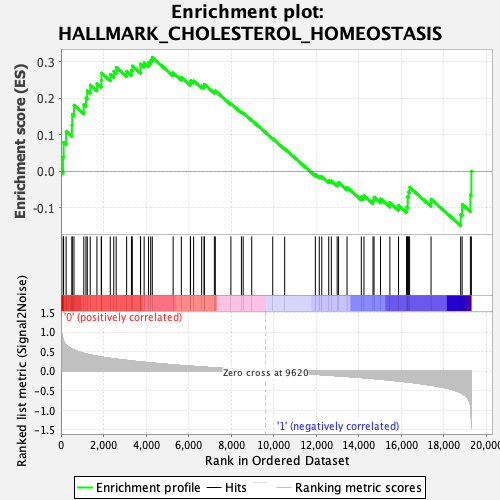
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.31212294 |
| Normalized Enrichment Score (NES) | 1.0312737 |
| Nominal p-value | 0.41260162 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.979 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mvk | 100 | 0.805 | 0.0389 | Yes |
| 2 | Fabp5 | 122 | 0.766 | 0.0797 | Yes |
| 3 | Tmem97 | 241 | 0.666 | 0.1100 | Yes |
| 4 | Mvd | 513 | 0.562 | 0.1267 | Yes |
| 5 | Trib3 | 529 | 0.558 | 0.1565 | Yes |
| 6 | Acat2 | 611 | 0.535 | 0.1816 | Yes |
| 7 | Lss | 1073 | 0.452 | 0.1823 | Yes |
| 8 | Abca2 | 1174 | 0.436 | 0.2010 | Yes |
| 9 | Hmgcs1 | 1237 | 0.427 | 0.2212 | Yes |
| 10 | Sqle | 1375 | 0.411 | 0.2365 | Yes |
| 11 | Dhcr7 | 1690 | 0.378 | 0.2409 | Yes |
| 12 | Antxr2 | 1895 | 0.360 | 0.2500 | Yes |
| 13 | Alcam | 1903 | 0.359 | 0.2693 | Yes |
| 14 | Fdps | 2317 | 0.325 | 0.2656 | Yes |
| 15 | Lpl | 2481 | 0.311 | 0.2741 | Yes |
| 16 | Srebf2 | 2595 | 0.305 | 0.2850 | Yes |
| 17 | Gnai1 | 3087 | 0.273 | 0.2744 | Yes |
| 18 | S100a11 | 3313 | 0.258 | 0.2768 | Yes |
| 19 | Fasn | 3355 | 0.256 | 0.2887 | Yes |
| 20 | Trp53inp1 | 3741 | 0.233 | 0.2814 | Yes |
| 21 | Hsd17b7 | 3742 | 0.233 | 0.2941 | Yes |
| 22 | Chka | 3910 | 0.225 | 0.2978 | Yes |
| 23 | Cyp51 | 4117 | 0.213 | 0.2987 | Yes |
| 24 | Hmgcr | 4219 | 0.207 | 0.3048 | Yes |
| 25 | Ctnnb1 | 4294 | 0.204 | 0.3121 | Yes |
| 26 | Jag1 | 5274 | 0.158 | 0.2699 | No |
| 27 | Fdft1 | 5660 | 0.142 | 0.2576 | No |
| 28 | Atxn2 | 6084 | 0.124 | 0.2424 | No |
| 29 | Cd9 | 6093 | 0.124 | 0.2488 | No |
| 30 | Fbxo6 | 6239 | 0.119 | 0.2478 | No |
| 31 | Pmvk | 6615 | 0.104 | 0.2340 | No |
| 32 | Pnrc1 | 6731 | 0.100 | 0.2335 | No |
| 33 | Plscr1 | 6741 | 0.099 | 0.2385 | No |
| 34 | Pparg | 7213 | 0.082 | 0.2184 | No |
| 35 | Tm7sf2 | 7258 | 0.080 | 0.2205 | No |
| 36 | Pdk3 | 7991 | 0.054 | 0.1855 | No |
| 37 | Cpeb2 | 8486 | 0.038 | 0.1619 | No |
| 38 | Stard4 | 8569 | 0.036 | 0.1596 | No |
| 39 | Fads2 | 8971 | 0.022 | 0.1399 | No |
| 40 | Ldlr | 9959 | -0.011 | 0.0892 | No |
| 41 | Gusb | 10521 | -0.029 | 0.0617 | No |
| 42 | Ethe1 | 11963 | -0.080 | -0.0088 | No |
| 43 | Clu | 12148 | -0.088 | -0.0136 | No |
| 44 | Stx5a | 12266 | -0.091 | -0.0147 | No |
| 45 | Idi1 | 12592 | -0.104 | -0.0259 | No |
| 46 | Gpx8 | 12715 | -0.109 | -0.0263 | No |
| 47 | Atf3 | 12989 | -0.119 | -0.0340 | No |
| 48 | Ech1 | 13056 | -0.120 | -0.0308 | No |
| 49 | Pcyt2 | 13453 | -0.136 | -0.0440 | No |
| 50 | Nfil3 | 14126 | -0.163 | -0.0700 | No |
| 51 | Ebp | 14250 | -0.169 | -0.0671 | No |
| 52 | Niban1 | 14675 | -0.188 | -0.0788 | No |
| 53 | Anxa5 | 14724 | -0.190 | -0.0709 | No |
| 54 | Lgals3 | 15028 | -0.206 | -0.0754 | No |
| 55 | Actg1 | 15469 | -0.228 | -0.0858 | No |
| 56 | Acss2 | 15876 | -0.249 | -0.0932 | No |
| 57 | Lgmn | 16251 | -0.273 | -0.0977 | No |
| 58 | Errfi1 | 16306 | -0.278 | -0.0853 | No |
| 59 | Nsdhl | 16307 | -0.278 | -0.0701 | No |
| 60 | Sc5d | 16346 | -0.280 | -0.0568 | No |
| 61 | Gldc | 16397 | -0.283 | -0.0439 | No |
| 62 | Atf5 | 17405 | -0.360 | -0.0765 | No |
| 63 | Aldoc | 18799 | -0.554 | -0.1186 | No |
| 64 | Gstm7 | 18868 | -0.574 | -0.0907 | No |
| 65 | Tnfrsf12a | 19255 | -0.844 | -0.0646 | No |
| 66 | Plaur | 19303 | -1.228 | 0.0002 | No |