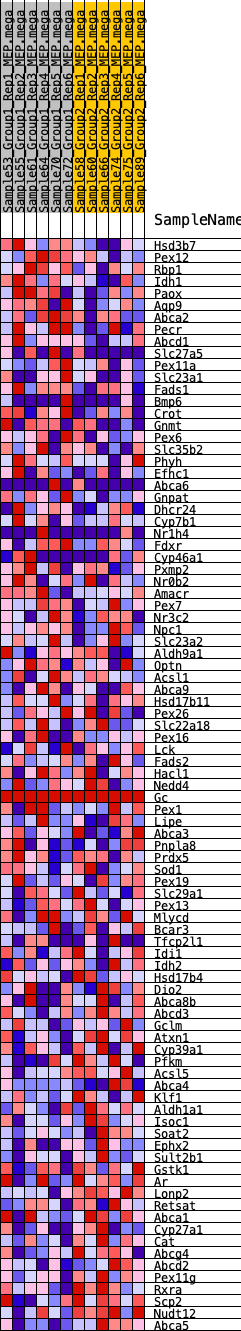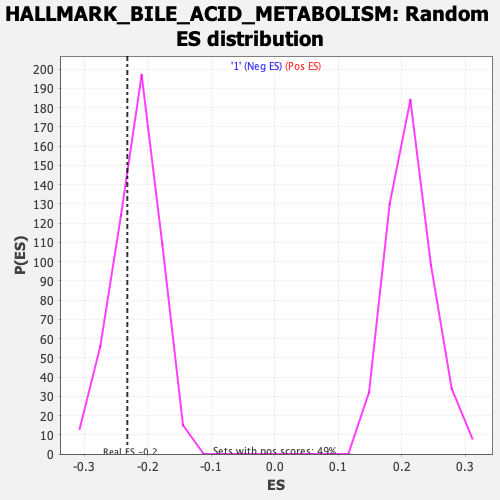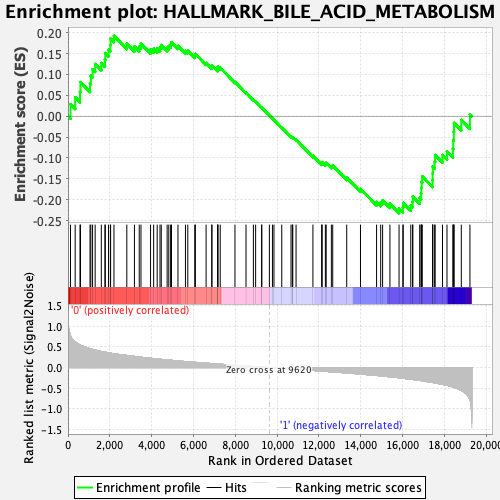
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.23224126 |
| Normalized Enrichment Score (NES) | -1.0625156 |
| Nominal p-value | 0.3171206 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.979 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd3b7 | 120 | 0.771 | 0.0287 | No |
| 2 | Pex12 | 343 | 0.620 | 0.0453 | No |
| 3 | Rbp1 | 574 | 0.549 | 0.0583 | No |
| 4 | Idh1 | 597 | 0.538 | 0.0815 | No |
| 5 | Paox | 1053 | 0.454 | 0.0784 | No |
| 6 | Aqp9 | 1092 | 0.450 | 0.0968 | No |
| 7 | Abca2 | 1174 | 0.436 | 0.1124 | No |
| 8 | Pecr | 1298 | 0.420 | 0.1250 | No |
| 9 | Abcd1 | 1592 | 0.386 | 0.1273 | No |
| 10 | Slc27a5 | 1766 | 0.372 | 0.1352 | No |
| 11 | Pex11a | 1783 | 0.370 | 0.1511 | No |
| 12 | Slc23a1 | 1938 | 0.355 | 0.1592 | No |
| 13 | Fads1 | 2026 | 0.347 | 0.1704 | No |
| 14 | Bmp6 | 2037 | 0.346 | 0.1856 | No |
| 15 | Crot | 2198 | 0.334 | 0.1924 | No |
| 16 | Gnmt | 2812 | 0.291 | 0.1737 | No |
| 17 | Pex6 | 3176 | 0.268 | 0.1669 | No |
| 18 | Slc35b2 | 3406 | 0.253 | 0.1665 | No |
| 19 | Phyh | 3489 | 0.248 | 0.1735 | No |
| 20 | Efhc1 | 3947 | 0.223 | 0.1598 | No |
| 21 | Abca6 | 4095 | 0.214 | 0.1619 | No |
| 22 | Gnpat | 4264 | 0.205 | 0.1624 | No |
| 23 | Dhcr24 | 4399 | 0.199 | 0.1645 | No |
| 24 | Cyp7b1 | 4459 | 0.197 | 0.1703 | No |
| 25 | Nr1h4 | 4740 | 0.183 | 0.1640 | No |
| 26 | Fdxr | 4819 | 0.178 | 0.1681 | No |
| 27 | Cyp46a1 | 4906 | 0.175 | 0.1715 | No |
| 28 | Pxmp2 | 4948 | 0.173 | 0.1772 | No |
| 29 | Nr0b2 | 5258 | 0.158 | 0.1683 | No |
| 30 | Amacr | 5615 | 0.143 | 0.1563 | No |
| 31 | Pex7 | 5727 | 0.138 | 0.1568 | No |
| 32 | Nr3c2 | 6061 | 0.125 | 0.1452 | No |
| 33 | Npc1 | 6090 | 0.124 | 0.1493 | No |
| 34 | Slc23a2 | 6602 | 0.105 | 0.1275 | No |
| 35 | Aldh9a1 | 6874 | 0.095 | 0.1177 | No |
| 36 | Optn | 6882 | 0.094 | 0.1216 | No |
| 37 | Acsl1 | 7154 | 0.084 | 0.1113 | No |
| 38 | Abca9 | 7164 | 0.084 | 0.1146 | No |
| 39 | Hsd17b11 | 7165 | 0.084 | 0.1185 | No |
| 40 | Pex26 | 7278 | 0.079 | 0.1162 | No |
| 41 | Slc22a18 | 7983 | 0.054 | 0.0821 | No |
| 42 | Pex16 | 8511 | 0.038 | 0.0563 | No |
| 43 | Lck | 8864 | 0.025 | 0.0392 | No |
| 44 | Fads2 | 8971 | 0.022 | 0.0346 | No |
| 45 | Hacl1 | 9258 | 0.012 | 0.0203 | No |
| 46 | Nedd4 | 9265 | 0.012 | 0.0205 | No |
| 47 | Gc | 9628 | 0.000 | 0.0017 | No |
| 48 | Pex1 | 9775 | -0.004 | -0.0057 | No |
| 49 | Lipe | 9847 | -0.006 | -0.0091 | No |
| 50 | Abca3 | 10220 | -0.019 | -0.0276 | No |
| 51 | Pnpla8 | 10663 | -0.034 | -0.0491 | No |
| 52 | Prdx5 | 10729 | -0.036 | -0.0508 | No |
| 53 | Sod1 | 10747 | -0.036 | -0.0501 | No |
| 54 | Pex19 | 10904 | -0.041 | -0.0563 | No |
| 55 | Slc29a1 | 11710 | -0.070 | -0.0950 | No |
| 56 | Pex13 | 12126 | -0.087 | -0.1127 | No |
| 57 | Mlycd | 12153 | -0.088 | -0.1101 | No |
| 58 | Bcar3 | 12306 | -0.092 | -0.1138 | No |
| 59 | Tfcp2l1 | 12344 | -0.094 | -0.1115 | No |
| 60 | Idi1 | 12592 | -0.104 | -0.1196 | No |
| 61 | Idh2 | 12657 | -0.107 | -0.1181 | No |
| 62 | Hsd17b4 | 13326 | -0.131 | -0.1469 | No |
| 63 | Dio2 | 13984 | -0.158 | -0.1739 | No |
| 64 | Abca8b | 14754 | -0.192 | -0.2053 | No |
| 65 | Abcd3 | 14950 | -0.202 | -0.2063 | No |
| 66 | Gclm | 15042 | -0.206 | -0.2016 | No |
| 67 | Atxn1 | 15392 | -0.225 | -0.2096 | No |
| 68 | Cyp39a1 | 15828 | -0.246 | -0.2211 | Yes |
| 69 | Pfkm | 16012 | -0.259 | -0.2189 | Yes |
| 70 | Acsl5 | 16025 | -0.259 | -0.2078 | Yes |
| 71 | Abca4 | 16389 | -0.282 | -0.2138 | Yes |
| 72 | Klf1 | 16476 | -0.288 | -0.2052 | Yes |
| 73 | Aldh1a1 | 16486 | -0.289 | -0.1926 | Yes |
| 74 | Isoc1 | 16815 | -0.310 | -0.1956 | Yes |
| 75 | Soat2 | 16881 | -0.315 | -0.1847 | Yes |
| 76 | Ephx2 | 16894 | -0.317 | -0.1710 | Yes |
| 77 | Sult2b1 | 16909 | -0.317 | -0.1573 | Yes |
| 78 | Gstk1 | 16936 | -0.321 | -0.1441 | Yes |
| 79 | Ar | 17429 | -0.361 | -0.1533 | Yes |
| 80 | Lonp2 | 17435 | -0.362 | -0.1372 | Yes |
| 81 | Retsat | 17442 | -0.363 | -0.1210 | Yes |
| 82 | Abca1 | 17531 | -0.369 | -0.1088 | Yes |
| 83 | Cyp27a1 | 17552 | -0.371 | -0.0930 | Yes |
| 84 | Cat | 17906 | -0.407 | -0.0930 | Yes |
| 85 | Abcg4 | 18120 | -0.429 | -0.0846 | Yes |
| 86 | Abcd2 | 18402 | -0.468 | -0.0780 | Yes |
| 87 | Pex11g | 18421 | -0.471 | -0.0576 | Yes |
| 88 | Rxra | 18446 | -0.476 | -0.0372 | Yes |
| 89 | Scp2 | 18458 | -0.477 | -0.0162 | Yes |
| 90 | Nudt12 | 18804 | -0.555 | -0.0090 | Yes |
| 91 | Abca5 | 19217 | -0.774 | 0.0047 | Yes |