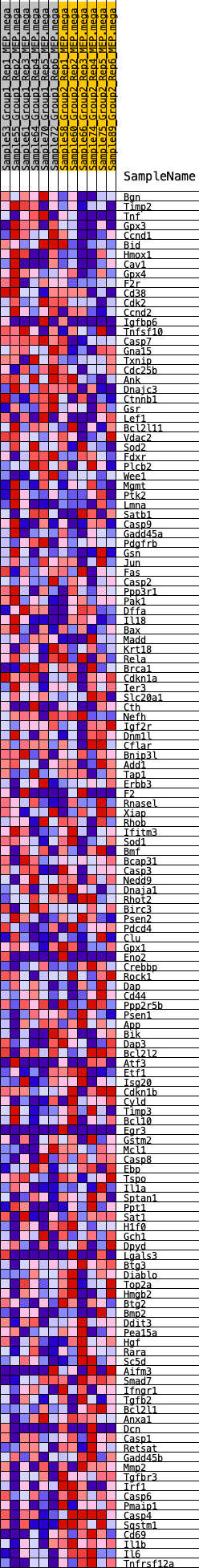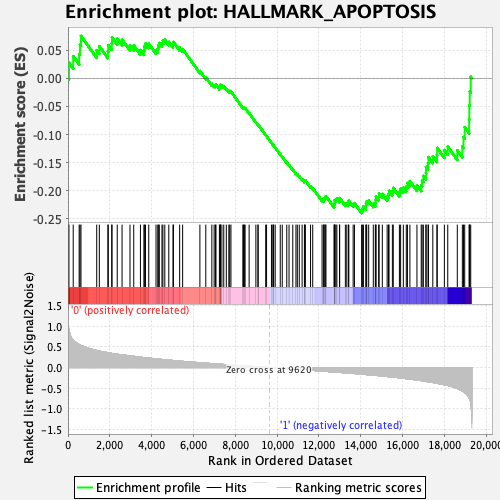
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.23984762 |
| Normalized Enrichment Score (NES) | -1.1219331 |
| Nominal p-value | 0.25101215 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.952 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bgn | 45 | 0.889 | 0.0276 | No |
| 2 | Timp2 | 249 | 0.662 | 0.0394 | No |
| 3 | Tnf | 533 | 0.557 | 0.0434 | No |
| 4 | Gpx3 | 573 | 0.549 | 0.0599 | No |
| 5 | Ccnd1 | 620 | 0.533 | 0.0755 | No |
| 6 | Bid | 1373 | 0.411 | 0.0501 | No |
| 7 | Hmox1 | 1498 | 0.398 | 0.0571 | No |
| 8 | Cav1 | 1907 | 0.359 | 0.0479 | No |
| 9 | Gpx4 | 1926 | 0.357 | 0.0590 | No |
| 10 | F2r | 2091 | 0.341 | 0.0620 | No |
| 11 | Cd38 | 2104 | 0.340 | 0.0728 | No |
| 12 | Cdk2 | 2354 | 0.322 | 0.0707 | No |
| 13 | Ccnd2 | 2585 | 0.305 | 0.0690 | No |
| 14 | Igfbp6 | 2964 | 0.280 | 0.0587 | No |
| 15 | Tnfsf10 | 3143 | 0.270 | 0.0585 | No |
| 16 | Casp7 | 3463 | 0.249 | 0.0503 | No |
| 17 | Gna15 | 3633 | 0.239 | 0.0496 | No |
| 18 | Txnip | 3649 | 0.238 | 0.0568 | No |
| 19 | Cdc25b | 3704 | 0.235 | 0.0619 | No |
| 20 | Ank | 3861 | 0.227 | 0.0614 | No |
| 21 | Dnajc3 | 4208 | 0.207 | 0.0504 | No |
| 22 | Ctnnb1 | 4294 | 0.204 | 0.0528 | No |
| 23 | Gsr | 4318 | 0.203 | 0.0585 | No |
| 24 | Lef1 | 4366 | 0.201 | 0.0628 | No |
| 25 | Bcl2l11 | 4493 | 0.195 | 0.0628 | No |
| 26 | Vdac2 | 4537 | 0.193 | 0.0670 | No |
| 27 | Sod2 | 4628 | 0.188 | 0.0687 | No |
| 28 | Fdxr | 4819 | 0.178 | 0.0648 | No |
| 29 | Plcb2 | 5020 | 0.169 | 0.0601 | No |
| 30 | Wee1 | 5041 | 0.168 | 0.0647 | No |
| 31 | Mgmt | 5338 | 0.154 | 0.0545 | No |
| 32 | Ptk2 | 5481 | 0.149 | 0.0521 | No |
| 33 | Lmna | 6306 | 0.117 | 0.0130 | No |
| 34 | Satb1 | 6588 | 0.105 | 0.0019 | No |
| 35 | Casp9 | 6878 | 0.094 | -0.0100 | No |
| 36 | Gadd45a | 6987 | 0.090 | -0.0126 | No |
| 37 | Pdgfrb | 7048 | 0.088 | -0.0127 | No |
| 38 | Gsn | 7075 | 0.087 | -0.0111 | No |
| 39 | Jun | 7239 | 0.081 | -0.0169 | No |
| 40 | Fas | 7275 | 0.079 | -0.0161 | No |
| 41 | Casp2 | 7283 | 0.079 | -0.0138 | No |
| 42 | Ppp3r1 | 7290 | 0.079 | -0.0114 | No |
| 43 | Pak1 | 7358 | 0.076 | -0.0123 | No |
| 44 | Dffa | 7447 | 0.073 | -0.0145 | No |
| 45 | Il18 | 7569 | 0.069 | -0.0184 | No |
| 46 | Bax | 7690 | 0.065 | -0.0225 | No |
| 47 | Madd | 7733 | 0.064 | -0.0225 | No |
| 48 | Krt18 | 7790 | 0.061 | -0.0234 | No |
| 49 | Rela | 8360 | 0.042 | -0.0517 | No |
| 50 | Brca1 | 8389 | 0.042 | -0.0517 | No |
| 51 | Cdkn1a | 8434 | 0.040 | -0.0527 | No |
| 52 | Ier3 | 8467 | 0.039 | -0.0530 | No |
| 53 | Slc20a1 | 8660 | 0.033 | -0.0619 | No |
| 54 | Cth | 8983 | 0.021 | -0.0780 | No |
| 55 | Nefh | 9085 | 0.018 | -0.0826 | No |
| 56 | Igf2r | 9088 | 0.018 | -0.0821 | No |
| 57 | Dnm1l | 9452 | 0.006 | -0.1009 | No |
| 58 | Cflar | 9494 | 0.005 | -0.1029 | No |
| 59 | Bnip3l | 9728 | -0.002 | -0.1149 | No |
| 60 | Add1 | 9800 | -0.005 | -0.1185 | No |
| 61 | Tap1 | 9806 | -0.005 | -0.1186 | No |
| 62 | Erbb3 | 9833 | -0.006 | -0.1198 | No |
| 63 | F2 | 9906 | -0.008 | -0.1232 | No |
| 64 | Rnasel | 10148 | -0.017 | -0.1352 | No |
| 65 | Xiap | 10244 | -0.020 | -0.1395 | No |
| 66 | Rhob | 10460 | -0.027 | -0.1498 | No |
| 67 | Ifitm3 | 10568 | -0.031 | -0.1544 | No |
| 68 | Sod1 | 10747 | -0.036 | -0.1624 | No |
| 69 | Bmf | 10899 | -0.041 | -0.1689 | No |
| 70 | Bcap31 | 10964 | -0.043 | -0.1708 | No |
| 71 | Casp3 | 11057 | -0.047 | -0.1740 | No |
| 72 | Nedd9 | 11193 | -0.052 | -0.1793 | No |
| 73 | Dnaja1 | 11303 | -0.055 | -0.1831 | No |
| 74 | Rhot2 | 11324 | -0.056 | -0.1823 | No |
| 75 | Birc3 | 11348 | -0.057 | -0.1815 | No |
| 76 | Psen2 | 11597 | -0.067 | -0.1922 | No |
| 77 | Pdcd4 | 11701 | -0.070 | -0.1953 | No |
| 78 | Clu | 12148 | -0.088 | -0.2156 | No |
| 79 | Gpx1 | 12226 | -0.090 | -0.2165 | No |
| 80 | Eno2 | 12230 | -0.090 | -0.2137 | No |
| 81 | Crebbp | 12282 | -0.091 | -0.2132 | No |
| 82 | Rock1 | 12298 | -0.092 | -0.2109 | No |
| 83 | Dap | 12334 | -0.094 | -0.2096 | No |
| 84 | Cd44 | 12726 | -0.109 | -0.2263 | No |
| 85 | Ppp2r5b | 12730 | -0.110 | -0.2228 | No |
| 86 | Psen1 | 12753 | -0.110 | -0.2202 | No |
| 87 | App | 12755 | -0.110 | -0.2165 | No |
| 88 | Bik | 12806 | -0.112 | -0.2154 | No |
| 89 | Dap3 | 12855 | -0.113 | -0.2140 | No |
| 90 | Bcl2l2 | 12983 | -0.118 | -0.2167 | No |
| 91 | Atf3 | 12989 | -0.119 | -0.2129 | No |
| 92 | Etf1 | 13261 | -0.127 | -0.2228 | No |
| 93 | Isg20 | 13329 | -0.131 | -0.2219 | No |
| 94 | Cdkn1b | 13407 | -0.134 | -0.2214 | No |
| 95 | Cyld | 13427 | -0.134 | -0.2178 | No |
| 96 | Timp3 | 13633 | -0.143 | -0.2237 | No |
| 97 | Bcl10 | 13696 | -0.145 | -0.2221 | No |
| 98 | Egr3 | 14038 | -0.161 | -0.2344 | Yes |
| 99 | Gstm2 | 14087 | -0.162 | -0.2315 | Yes |
| 100 | Mcl1 | 14124 | -0.163 | -0.2278 | Yes |
| 101 | Casp8 | 14247 | -0.169 | -0.2285 | Yes |
| 102 | Ebp | 14250 | -0.169 | -0.2229 | Yes |
| 103 | Tspo | 14283 | -0.171 | -0.2188 | Yes |
| 104 | Il1a | 14376 | -0.176 | -0.2177 | Yes |
| 105 | Sptan1 | 14597 | -0.184 | -0.2229 | Yes |
| 106 | Ppt1 | 14693 | -0.189 | -0.2215 | Yes |
| 107 | Sat1 | 14716 | -0.190 | -0.2163 | Yes |
| 108 | H1f0 | 14725 | -0.190 | -0.2103 | Yes |
| 109 | Gch1 | 14855 | -0.196 | -0.2104 | Yes |
| 110 | Dpyd | 14873 | -0.197 | -0.2046 | Yes |
| 111 | Lgals3 | 15028 | -0.206 | -0.2057 | Yes |
| 112 | Btg3 | 15259 | -0.217 | -0.2104 | Yes |
| 113 | Diablo | 15322 | -0.220 | -0.2062 | Yes |
| 114 | Top2a | 15350 | -0.222 | -0.2001 | Yes |
| 115 | Hmgb2 | 15509 | -0.230 | -0.2006 | Yes |
| 116 | Btg2 | 15551 | -0.232 | -0.1949 | Yes |
| 117 | Bmp2 | 15845 | -0.247 | -0.2018 | Yes |
| 118 | Ddit3 | 15904 | -0.251 | -0.1964 | Yes |
| 119 | Pea15a | 16033 | -0.260 | -0.1943 | Yes |
| 120 | Hgf | 16174 | -0.269 | -0.1925 | Yes |
| 121 | Rara | 16229 | -0.272 | -0.1862 | Yes |
| 122 | Sc5d | 16346 | -0.280 | -0.1828 | Yes |
| 123 | Aifm3 | 16684 | -0.303 | -0.1901 | Yes |
| 124 | Smad7 | 16888 | -0.316 | -0.1901 | Yes |
| 125 | Ifngr1 | 16931 | -0.321 | -0.1814 | Yes |
| 126 | Tgfb2 | 16990 | -0.325 | -0.1735 | Yes |
| 127 | Bcl2l1 | 17114 | -0.335 | -0.1686 | Yes |
| 128 | Anxa1 | 17120 | -0.336 | -0.1576 | Yes |
| 129 | Dcn | 17214 | -0.344 | -0.1508 | Yes |
| 130 | Casp1 | 17234 | -0.346 | -0.1401 | Yes |
| 131 | Retsat | 17442 | -0.363 | -0.1387 | Yes |
| 132 | Gadd45b | 17641 | -0.380 | -0.1362 | Yes |
| 133 | Mmp2 | 17651 | -0.381 | -0.1238 | Yes |
| 134 | Tgfbr3 | 17995 | -0.416 | -0.1277 | Yes |
| 135 | Irf1 | 18157 | -0.434 | -0.1214 | Yes |
| 136 | Casp6 | 18613 | -0.509 | -0.1280 | Yes |
| 137 | Pmaip1 | 18854 | -0.571 | -0.1212 | Yes |
| 138 | Casp4 | 18909 | -0.583 | -0.1044 | Yes |
| 139 | Sqstm1 | 18967 | -0.605 | -0.0869 | Yes |
| 140 | Cd69 | 19181 | -0.739 | -0.0731 | Yes |
| 141 | Il1b | 19189 | -0.746 | -0.0483 | Yes |
| 142 | Il6 | 19208 | -0.767 | -0.0234 | Yes |
| 143 | Tnfrsf12a | 19255 | -0.844 | 0.0027 | Yes |