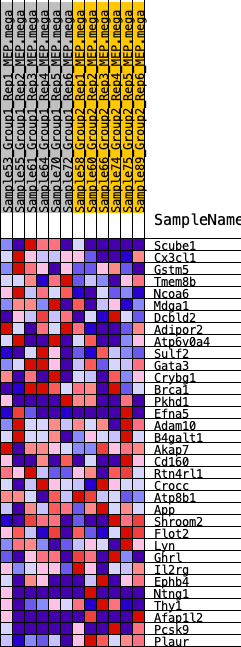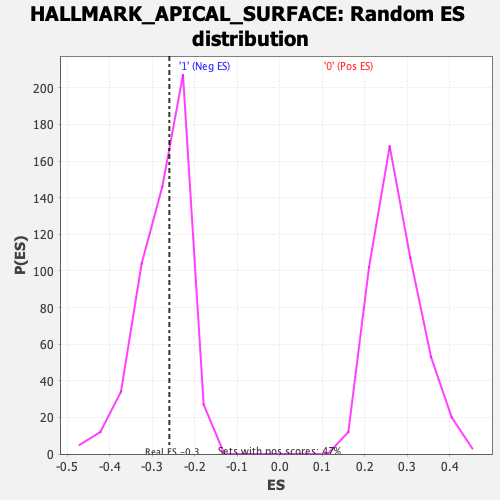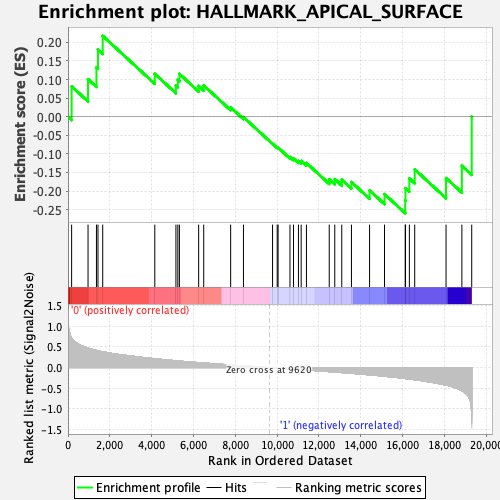
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_SURFACE |
| Enrichment Score (ES) | -0.25954387 |
| Normalized Enrichment Score (NES) | -0.9497028 |
| Nominal p-value | 0.5102804 |
| FDR q-value | 0.96603215 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Scube1 | 173 | 0.710 | 0.0817 | No |
| 2 | Cx3cl1 | 958 | 0.470 | 0.1010 | No |
| 3 | Gstm5 | 1360 | 0.412 | 0.1328 | No |
| 4 | Tmem8b | 1433 | 0.404 | 0.1807 | No |
| 5 | Ncoa6 | 1660 | 0.381 | 0.2176 | No |
| 6 | Mdga1 | 4150 | 0.211 | 0.1154 | No |
| 7 | Dcbld2 | 5160 | 0.162 | 0.0838 | No |
| 8 | Adipor2 | 5252 | 0.158 | 0.0993 | No |
| 9 | Atp6v0a4 | 5326 | 0.155 | 0.1153 | No |
| 10 | Sulf2 | 6247 | 0.119 | 0.0828 | No |
| 11 | Gata3 | 6489 | 0.109 | 0.0842 | No |
| 12 | Crybg1 | 7777 | 0.062 | 0.0254 | No |
| 13 | Brca1 | 8389 | 0.042 | -0.0010 | No |
| 14 | Pkhd1 | 9780 | -0.004 | -0.0726 | No |
| 15 | Efna5 | 10002 | -0.012 | -0.0826 | No |
| 16 | Adam10 | 10045 | -0.013 | -0.0831 | No |
| 17 | B4galt1 | 10613 | -0.032 | -0.1084 | No |
| 18 | Akap7 | 10782 | -0.038 | -0.1123 | No |
| 19 | Cd160 | 11018 | -0.045 | -0.1187 | No |
| 20 | Rtn4rl1 | 11148 | -0.050 | -0.1190 | No |
| 21 | Crocc | 11400 | -0.059 | -0.1245 | No |
| 22 | Atp8b1 | 12490 | -0.099 | -0.1683 | No |
| 23 | App | 12755 | -0.110 | -0.1679 | No |
| 24 | Shroom2 | 13089 | -0.121 | -0.1697 | No |
| 25 | Flot2 | 13552 | -0.139 | -0.1759 | No |
| 26 | Lyn | 14419 | -0.177 | -0.1982 | No |
| 27 | Ghrl | 15133 | -0.211 | -0.2083 | No |
| 28 | Il2rg | 16122 | -0.266 | -0.2256 | Yes |
| 29 | Ephb4 | 16137 | -0.267 | -0.1923 | Yes |
| 30 | Ntng1 | 16321 | -0.278 | -0.1662 | Yes |
| 31 | Thy1 | 16577 | -0.295 | -0.1417 | Yes |
| 32 | Afap1l2 | 18075 | -0.425 | -0.1651 | Yes |
| 33 | Pcsk9 | 18832 | -0.564 | -0.1323 | Yes |
| 34 | Plaur | 19303 | -1.228 | 0.0002 | Yes |