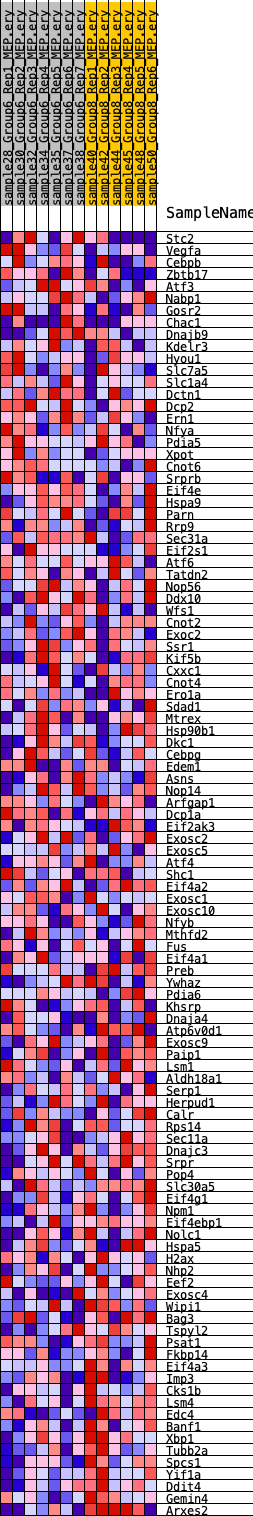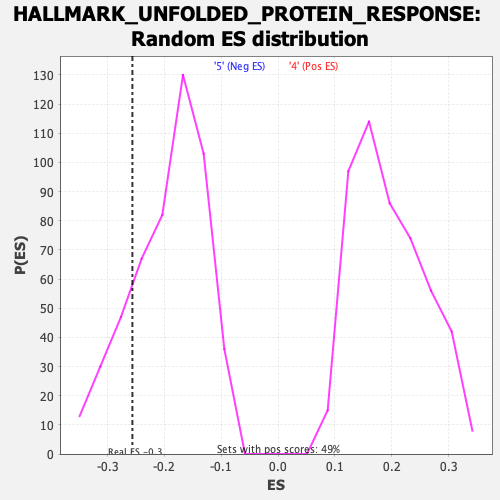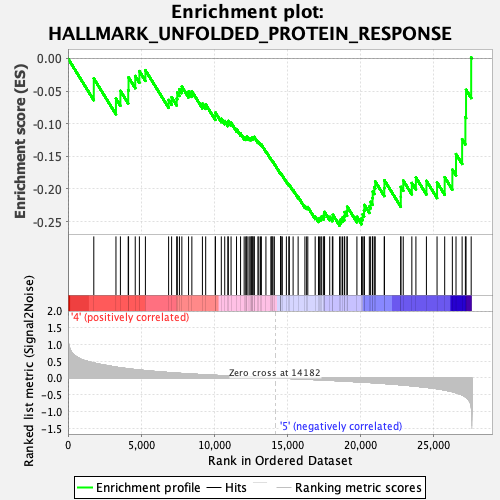
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group6_versus_Group8.MEP.ery_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.ery_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.2562474 |
| Normalized Enrichment Score (NES) | -1.3302237 |
| Nominal p-value | 0.18307087 |
| FDR q-value | 0.38212508 |
| FWER p-Value | 0.718 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 1763 | 0.445 | -0.0307 | No |
| 2 | Vegfa | 3278 | 0.323 | -0.0615 | No |
| 3 | Cebpb | 3582 | 0.303 | -0.0497 | No |
| 4 | Zbtb17 | 4115 | 0.272 | -0.0486 | No |
| 5 | Atf3 | 4139 | 0.272 | -0.0290 | No |
| 6 | Nabp1 | 4596 | 0.248 | -0.0269 | No |
| 7 | Gosr2 | 4880 | 0.236 | -0.0195 | No |
| 8 | Chac1 | 5291 | 0.217 | -0.0181 | No |
| 9 | Dnajb9 | 6877 | 0.160 | -0.0637 | No |
| 10 | Kdelr3 | 7082 | 0.154 | -0.0595 | No |
| 11 | Hyou1 | 7431 | 0.143 | -0.0614 | No |
| 12 | Slc7a5 | 7469 | 0.142 | -0.0521 | No |
| 13 | Slc1a4 | 7617 | 0.137 | -0.0472 | No |
| 14 | Dctn1 | 7782 | 0.132 | -0.0432 | No |
| 15 | Dcp2 | 8240 | 0.121 | -0.0507 | No |
| 16 | Ern1 | 8466 | 0.115 | -0.0503 | No |
| 17 | Nfya | 9188 | 0.097 | -0.0692 | No |
| 18 | Pdia5 | 9411 | 0.092 | -0.0704 | No |
| 19 | Xpot | 10067 | 0.077 | -0.0885 | No |
| 20 | Cnot6 | 10075 | 0.077 | -0.0829 | No |
| 21 | Srprb | 10479 | 0.069 | -0.0924 | No |
| 22 | Eif4e | 10713 | 0.065 | -0.0960 | No |
| 23 | Hspa9 | 10927 | 0.061 | -0.0992 | No |
| 24 | Parn | 10957 | 0.060 | -0.0958 | No |
| 25 | Rrp9 | 11146 | 0.056 | -0.0984 | No |
| 26 | Sec31a | 11519 | 0.049 | -0.1083 | No |
| 27 | Eif2s1 | 11795 | 0.044 | -0.1150 | No |
| 28 | Atf6 | 12041 | 0.039 | -0.1210 | No |
| 29 | Tatdn2 | 12149 | 0.037 | -0.1221 | No |
| 30 | Nop56 | 12221 | 0.035 | -0.1220 | No |
| 31 | Ddx10 | 12232 | 0.035 | -0.1197 | No |
| 32 | Wfs1 | 12372 | 0.032 | -0.1224 | No |
| 33 | Cnot2 | 12448 | 0.031 | -0.1228 | No |
| 34 | Exoc2 | 12519 | 0.030 | -0.1231 | No |
| 35 | Ssr1 | 12568 | 0.029 | -0.1227 | No |
| 36 | Kif5b | 12587 | 0.028 | -0.1212 | No |
| 37 | Cxxc1 | 12638 | 0.027 | -0.1210 | No |
| 38 | Cnot4 | 12717 | 0.026 | -0.1218 | No |
| 39 | Ero1a | 12729 | 0.026 | -0.1203 | No |
| 40 | Sdad1 | 12982 | 0.022 | -0.1278 | No |
| 41 | Mtrex | 13046 | 0.021 | -0.1285 | No |
| 42 | Hsp90b1 | 13162 | 0.018 | -0.1313 | No |
| 43 | Dkc1 | 13217 | 0.017 | -0.1320 | No |
| 44 | Cebpg | 13529 | 0.012 | -0.1424 | No |
| 45 | Edem1 | 13867 | 0.005 | -0.1543 | No |
| 46 | Asns | 13931 | 0.004 | -0.1562 | No |
| 47 | Nop14 | 13950 | 0.004 | -0.1566 | No |
| 48 | Arfgap1 | 14011 | 0.003 | -0.1585 | No |
| 49 | Dcp1a | 14105 | 0.001 | -0.1618 | No |
| 50 | Eif2ak3 | 14529 | -0.004 | -0.1769 | No |
| 51 | Exosc2 | 14532 | -0.004 | -0.1766 | No |
| 52 | Exosc5 | 14546 | -0.005 | -0.1767 | No |
| 53 | Atf4 | 14559 | -0.005 | -0.1768 | No |
| 54 | Shc1 | 14649 | -0.006 | -0.1796 | No |
| 55 | Eif4a2 | 14927 | -0.011 | -0.1888 | No |
| 56 | Exosc1 | 15095 | -0.014 | -0.1938 | No |
| 57 | Exosc10 | 15128 | -0.015 | -0.1939 | No |
| 58 | Nfyb | 15387 | -0.020 | -0.2018 | No |
| 59 | Mthfd2 | 15730 | -0.026 | -0.2122 | No |
| 60 | Fus | 16194 | -0.035 | -0.2265 | No |
| 61 | Eif4a1 | 16311 | -0.037 | -0.2279 | No |
| 62 | Preb | 16392 | -0.039 | -0.2279 | No |
| 63 | Ywhaz | 16895 | -0.048 | -0.2426 | No |
| 64 | Pdia6 | 17121 | -0.053 | -0.2467 | No |
| 65 | Khsrp | 17183 | -0.054 | -0.2449 | No |
| 66 | Dnaja4 | 17282 | -0.056 | -0.2442 | No |
| 67 | Atp6v0d1 | 17344 | -0.058 | -0.2421 | No |
| 68 | Exosc9 | 17478 | -0.060 | -0.2424 | No |
| 69 | Paip1 | 17506 | -0.061 | -0.2388 | No |
| 70 | Lsm1 | 17537 | -0.061 | -0.2353 | No |
| 71 | Aldh18a1 | 17890 | -0.069 | -0.2429 | No |
| 72 | Serp1 | 18070 | -0.073 | -0.2439 | No |
| 73 | Herpud1 | 18106 | -0.074 | -0.2397 | No |
| 74 | Calr | 18563 | -0.085 | -0.2499 | Yes |
| 75 | Rps14 | 18653 | -0.087 | -0.2466 | Yes |
| 76 | Sec11a | 18761 | -0.089 | -0.2438 | Yes |
| 77 | Dnajc3 | 18883 | -0.091 | -0.2414 | Yes |
| 78 | Srpr | 18913 | -0.092 | -0.2355 | Yes |
| 79 | Pop4 | 19072 | -0.096 | -0.2340 | Yes |
| 80 | Slc30a5 | 19084 | -0.097 | -0.2271 | Yes |
| 81 | Eif4g1 | 19748 | -0.113 | -0.2428 | Yes |
| 82 | Npm1 | 20063 | -0.119 | -0.2452 | Yes |
| 83 | Eif4ebp1 | 20143 | -0.121 | -0.2390 | Yes |
| 84 | Nolc1 | 20236 | -0.123 | -0.2331 | Yes |
| 85 | Hspa5 | 20259 | -0.124 | -0.2246 | Yes |
| 86 | H2ax | 20598 | -0.133 | -0.2269 | Yes |
| 87 | Nhp2 | 20686 | -0.136 | -0.2198 | Yes |
| 88 | Eef2 | 20813 | -0.139 | -0.2139 | Yes |
| 89 | Exosc4 | 20830 | -0.140 | -0.2040 | Yes |
| 90 | Wipi1 | 20937 | -0.143 | -0.1972 | Yes |
| 91 | Bag3 | 21000 | -0.144 | -0.1886 | Yes |
| 92 | Tspyl2 | 21617 | -0.163 | -0.1987 | Yes |
| 93 | Psat1 | 21630 | -0.164 | -0.1868 | Yes |
| 94 | Fkbp14 | 22748 | -0.204 | -0.2122 | Yes |
| 95 | Eif4a3 | 22751 | -0.204 | -0.1969 | Yes |
| 96 | Imp3 | 22915 | -0.209 | -0.1871 | Yes |
| 97 | Cks1b | 23503 | -0.233 | -0.1909 | Yes |
| 98 | Lsm4 | 23781 | -0.245 | -0.1826 | Yes |
| 99 | Edc4 | 24499 | -0.279 | -0.1877 | Yes |
| 100 | Banf1 | 25226 | -0.319 | -0.1901 | Yes |
| 101 | Xbp1 | 25748 | -0.358 | -0.1821 | Yes |
| 102 | Tubb2a | 26272 | -0.408 | -0.1705 | Yes |
| 103 | Spcs1 | 26522 | -0.438 | -0.1466 | Yes |
| 104 | Yif1a | 26940 | -0.505 | -0.1239 | Yes |
| 105 | Ddit4 | 27163 | -0.560 | -0.0898 | Yes |
| 106 | Gemin4 | 27208 | -0.577 | -0.0480 | Yes |
| 107 | Arxes2 | 27559 | -0.830 | 0.0016 | Yes |