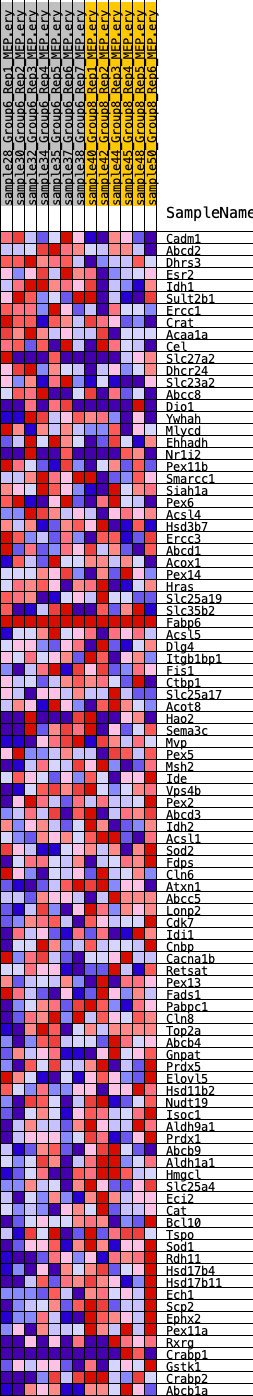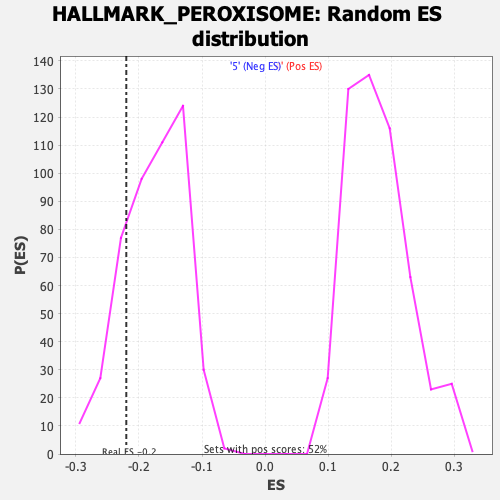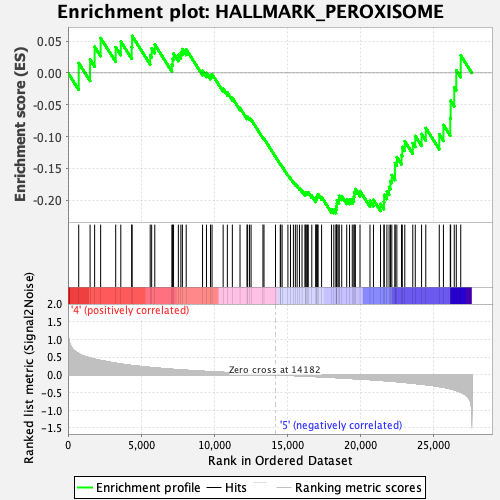
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group6_versus_Group8.MEP.ery_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.ery_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.22009636 |
| Normalized Enrichment Score (NES) | -1.2457516 |
| Nominal p-value | 0.19791667 |
| FDR q-value | 0.38717645 |
| FWER p-Value | 0.8 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cadm1 | 732 | 0.592 | 0.0155 | No |
| 2 | Abcd2 | 1509 | 0.473 | 0.0210 | No |
| 3 | Dhrs3 | 1820 | 0.439 | 0.0410 | No |
| 4 | Esr2 | 2231 | 0.401 | 0.0547 | No |
| 5 | Idh1 | 3260 | 0.325 | 0.0404 | No |
| 6 | Sult2b1 | 3609 | 0.302 | 0.0493 | No |
| 7 | Ercc1 | 4352 | 0.260 | 0.0408 | No |
| 8 | Crat | 4383 | 0.258 | 0.0581 | No |
| 9 | Acaa1a | 5618 | 0.205 | 0.0278 | No |
| 10 | Cel | 5719 | 0.200 | 0.0385 | No |
| 11 | Slc27a2 | 5932 | 0.193 | 0.0445 | No |
| 12 | Dhcr24 | 7101 | 0.154 | 0.0130 | No |
| 13 | Slc23a2 | 7147 | 0.152 | 0.0222 | No |
| 14 | Abcc8 | 7215 | 0.150 | 0.0304 | No |
| 15 | Dio1 | 7543 | 0.140 | 0.0284 | No |
| 16 | Ywhah | 7715 | 0.135 | 0.0318 | No |
| 17 | Mlycd | 7819 | 0.131 | 0.0374 | No |
| 18 | Ehhadh | 8079 | 0.125 | 0.0369 | No |
| 19 | Nr1i2 | 9197 | 0.097 | 0.0032 | No |
| 20 | Pex11b | 9465 | 0.091 | -0.0001 | No |
| 21 | Smarcc1 | 9739 | 0.085 | -0.0040 | No |
| 22 | Siah1a | 9845 | 0.082 | -0.0019 | No |
| 23 | Pex6 | 10605 | 0.066 | -0.0248 | No |
| 24 | Acsl4 | 10892 | 0.061 | -0.0308 | No |
| 25 | Hsd3b7 | 11234 | 0.054 | -0.0393 | No |
| 26 | Ercc3 | 11758 | 0.044 | -0.0552 | No |
| 27 | Abcd1 | 12235 | 0.035 | -0.0700 | No |
| 28 | Acox1 | 12279 | 0.034 | -0.0691 | No |
| 29 | Pex14 | 12410 | 0.032 | -0.0715 | No |
| 30 | Hras | 12514 | 0.030 | -0.0732 | No |
| 31 | Slc25a19 | 13325 | 0.015 | -0.1015 | No |
| 32 | Slc35b2 | 13397 | 0.014 | -0.1031 | No |
| 33 | Fabp6 | 14182 | 0.000 | -0.1316 | No |
| 34 | Acsl5 | 14509 | -0.004 | -0.1432 | No |
| 35 | Dlg4 | 14540 | -0.005 | -0.1439 | No |
| 36 | Itgb1bp1 | 14637 | -0.006 | -0.1470 | No |
| 37 | Fis1 | 15032 | -0.013 | -0.1604 | No |
| 38 | Ctbp1 | 15208 | -0.016 | -0.1656 | No |
| 39 | Slc25a17 | 15407 | -0.020 | -0.1714 | No |
| 40 | Acot8 | 15541 | -0.022 | -0.1746 | No |
| 41 | Hao2 | 15664 | -0.025 | -0.1773 | No |
| 42 | Sema3c | 15822 | -0.028 | -0.1810 | No |
| 43 | Mvp | 15996 | -0.031 | -0.1851 | No |
| 44 | Pex5 | 16208 | -0.035 | -0.1903 | No |
| 45 | Msh2 | 16212 | -0.035 | -0.1879 | No |
| 46 | Ide | 16290 | -0.037 | -0.1881 | No |
| 47 | Vps4b | 16377 | -0.038 | -0.1885 | No |
| 48 | Pex2 | 16420 | -0.039 | -0.1872 | No |
| 49 | Abcd3 | 16664 | -0.044 | -0.1929 | No |
| 50 | Idh2 | 16933 | -0.049 | -0.1991 | No |
| 51 | Acsl1 | 16960 | -0.050 | -0.1965 | No |
| 52 | Sod2 | 17007 | -0.051 | -0.1946 | No |
| 53 | Fdps | 17045 | -0.052 | -0.1923 | No |
| 54 | Cln6 | 17096 | -0.053 | -0.1903 | No |
| 55 | Atxn1 | 17334 | -0.057 | -0.1949 | No |
| 56 | Abcc5 | 18012 | -0.072 | -0.2144 | No |
| 57 | Lonp2 | 18171 | -0.075 | -0.2147 | Yes |
| 58 | Cdk7 | 18319 | -0.078 | -0.2145 | Yes |
| 59 | Idi1 | 18334 | -0.079 | -0.2094 | Yes |
| 60 | Cnbp | 18376 | -0.080 | -0.2052 | Yes |
| 61 | Cacna1b | 18385 | -0.080 | -0.1998 | Yes |
| 62 | Retsat | 18522 | -0.084 | -0.1987 | Yes |
| 63 | Pex13 | 18528 | -0.084 | -0.1929 | Yes |
| 64 | Fads1 | 18709 | -0.087 | -0.1933 | Yes |
| 65 | Pabpc1 | 19050 | -0.096 | -0.1988 | Yes |
| 66 | Cln8 | 19244 | -0.100 | -0.1987 | Yes |
| 67 | Top2a | 19431 | -0.105 | -0.1980 | Yes |
| 68 | Abcb4 | 19545 | -0.108 | -0.1944 | Yes |
| 69 | Gnpat | 19562 | -0.108 | -0.1873 | Yes |
| 70 | Prdx5 | 19643 | -0.110 | -0.1823 | Yes |
| 71 | Elovl5 | 19960 | -0.117 | -0.1855 | Yes |
| 72 | Hsd11b2 | 20642 | -0.134 | -0.2006 | Yes |
| 73 | Nudt19 | 20880 | -0.141 | -0.1992 | Yes |
| 74 | Isoc1 | 21362 | -0.156 | -0.2056 | Yes |
| 75 | Aldh9a1 | 21595 | -0.163 | -0.2024 | Yes |
| 76 | Prdx1 | 21613 | -0.163 | -0.1914 | Yes |
| 77 | Abcb9 | 21798 | -0.170 | -0.1860 | Yes |
| 78 | Aldh1a1 | 21948 | -0.175 | -0.1790 | Yes |
| 79 | Hmgcl | 22041 | -0.176 | -0.1697 | Yes |
| 80 | Slc25a4 | 22134 | -0.180 | -0.1603 | Yes |
| 81 | Eci2 | 22353 | -0.188 | -0.1548 | Yes |
| 82 | Cat | 22355 | -0.188 | -0.1415 | Yes |
| 83 | Bcl10 | 22479 | -0.193 | -0.1322 | Yes |
| 84 | Tspo | 22796 | -0.205 | -0.1291 | Yes |
| 85 | Sod1 | 22849 | -0.207 | -0.1162 | Yes |
| 86 | Rdh11 | 23021 | -0.213 | -0.1072 | Yes |
| 87 | Hsd17b4 | 23567 | -0.236 | -0.1102 | Yes |
| 88 | Hsd17b11 | 23737 | -0.244 | -0.0989 | Yes |
| 89 | Ech1 | 24168 | -0.263 | -0.0958 | Yes |
| 90 | Scp2 | 24456 | -0.278 | -0.0865 | Yes |
| 91 | Ephx2 | 25378 | -0.330 | -0.0964 | Yes |
| 92 | Pex11a | 25662 | -0.351 | -0.0817 | Yes |
| 93 | Rxrg | 26130 | -0.392 | -0.0708 | Yes |
| 94 | Crabp1 | 26156 | -0.395 | -0.0436 | Yes |
| 95 | Gstk1 | 26402 | -0.422 | -0.0224 | Yes |
| 96 | Crabp2 | 26541 | -0.441 | 0.0040 | Yes |
| 97 | Abcb1a | 26845 | -0.485 | 0.0275 | Yes |