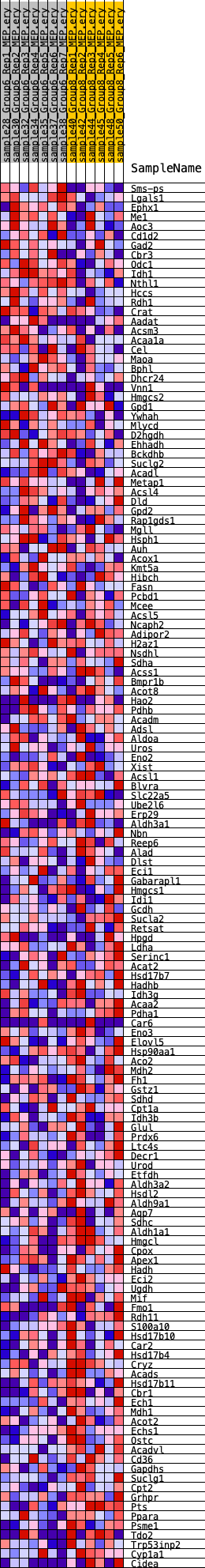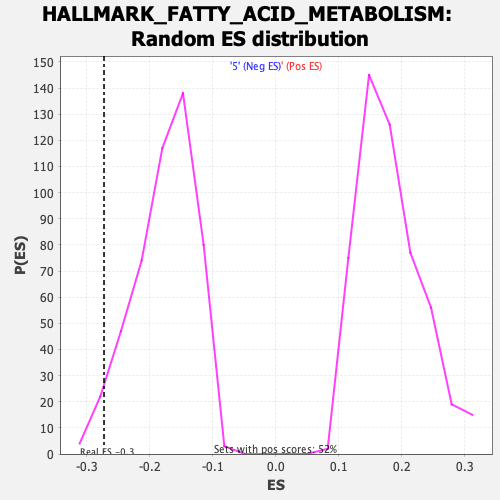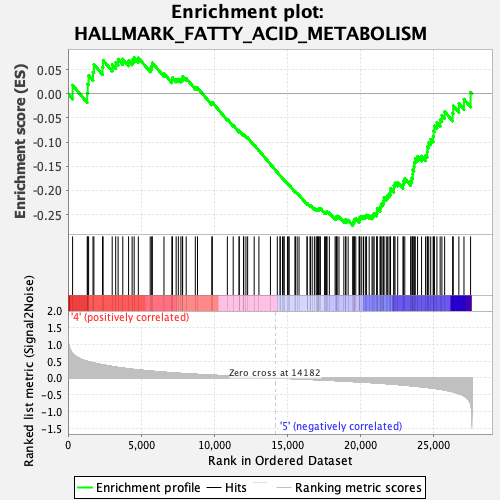
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group6_versus_Group8.MEP.ery_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.ery_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.27274606 |
| Normalized Enrichment Score (NES) | -1.5436089 |
| Nominal p-value | 0.035051547 |
| FDR q-value | 0.21384303 |
| FWER p-Value | 0.398 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sms-ps | 314 | 0.724 | 0.0178 | No |
| 2 | Lgals1 | 1314 | 0.498 | 0.0015 | No |
| 3 | Ephx1 | 1348 | 0.493 | 0.0202 | No |
| 4 | Me1 | 1408 | 0.486 | 0.0377 | No |
| 5 | Aoc3 | 1709 | 0.451 | 0.0449 | No |
| 6 | Cd1d2 | 1770 | 0.444 | 0.0607 | No |
| 7 | Gad2 | 2359 | 0.391 | 0.0550 | No |
| 8 | Cbr3 | 2399 | 0.388 | 0.0693 | No |
| 9 | Odc1 | 3021 | 0.341 | 0.0605 | No |
| 10 | Idh1 | 3260 | 0.325 | 0.0649 | No |
| 11 | Nthl1 | 3425 | 0.313 | 0.0715 | No |
| 12 | Hccs | 3745 | 0.293 | 0.0717 | No |
| 13 | Rdh1 | 4140 | 0.272 | 0.0684 | No |
| 14 | Crat | 4383 | 0.258 | 0.0700 | No |
| 15 | Aadat | 4522 | 0.251 | 0.0751 | No |
| 16 | Acsm3 | 4807 | 0.239 | 0.0744 | No |
| 17 | Acaa1a | 5618 | 0.205 | 0.0532 | No |
| 18 | Cel | 5719 | 0.200 | 0.0576 | No |
| 19 | Maoa | 5765 | 0.200 | 0.0640 | No |
| 20 | Bphl | 6560 | 0.170 | 0.0420 | No |
| 21 | Dhcr24 | 7101 | 0.154 | 0.0285 | No |
| 22 | Vnn1 | 7140 | 0.152 | 0.0333 | No |
| 23 | Hmgcs2 | 7392 | 0.144 | 0.0300 | No |
| 24 | Gpd1 | 7542 | 0.140 | 0.0302 | No |
| 25 | Ywhah | 7715 | 0.135 | 0.0293 | No |
| 26 | Mlycd | 7819 | 0.131 | 0.0309 | No |
| 27 | D2hgdh | 7835 | 0.131 | 0.0356 | No |
| 28 | Ehhadh | 8079 | 0.125 | 0.0318 | No |
| 29 | Bckdhb | 8702 | 0.109 | 0.0135 | No |
| 30 | Suclg2 | 8851 | 0.105 | 0.0124 | No |
| 31 | Acadl | 9831 | 0.083 | -0.0199 | No |
| 32 | Metap1 | 9864 | 0.082 | -0.0178 | No |
| 33 | Acsl4 | 10892 | 0.061 | -0.0527 | No |
| 34 | Dld | 11292 | 0.053 | -0.0651 | No |
| 35 | Gpd2 | 11694 | 0.046 | -0.0778 | No |
| 36 | Rap1gds1 | 11701 | 0.046 | -0.0762 | No |
| 37 | Mgll | 11998 | 0.040 | -0.0854 | No |
| 38 | Hsph1 | 12013 | 0.040 | -0.0843 | No |
| 39 | Auh | 12163 | 0.037 | -0.0883 | No |
| 40 | Acox1 | 12279 | 0.034 | -0.0911 | No |
| 41 | Kmt5a | 12726 | 0.026 | -0.1062 | No |
| 42 | Hibch | 13050 | 0.021 | -0.1172 | No |
| 43 | Fasn | 13837 | 0.006 | -0.1456 | No |
| 44 | Pcbd1 | 14307 | -0.000 | -0.1626 | No |
| 45 | Mcee | 14491 | -0.004 | -0.1691 | No |
| 46 | Acsl5 | 14509 | -0.004 | -0.1696 | No |
| 47 | Ncaph2 | 14662 | -0.006 | -0.1749 | No |
| 48 | Adipor2 | 14757 | -0.008 | -0.1780 | No |
| 49 | H2az1 | 14770 | -0.008 | -0.1781 | No |
| 50 | Nsdhl | 15000 | -0.012 | -0.1859 | No |
| 51 | Sdha | 15055 | -0.013 | -0.1873 | No |
| 52 | Acss1 | 15118 | -0.015 | -0.1890 | No |
| 53 | Bmpr1b | 15515 | -0.022 | -0.2026 | No |
| 54 | Acot8 | 15541 | -0.022 | -0.2026 | No |
| 55 | Hao2 | 15664 | -0.025 | -0.2060 | No |
| 56 | Pdhb | 15792 | -0.027 | -0.2095 | No |
| 57 | Acadm | 16341 | -0.037 | -0.2280 | No |
| 58 | Adsl | 16346 | -0.038 | -0.2266 | No |
| 59 | Aldoa | 16545 | -0.042 | -0.2321 | No |
| 60 | Uros | 16574 | -0.042 | -0.2314 | No |
| 61 | Eno2 | 16692 | -0.044 | -0.2339 | No |
| 62 | Xist | 16841 | -0.047 | -0.2374 | No |
| 63 | Acsl1 | 16960 | -0.050 | -0.2397 | No |
| 64 | Blvra | 17021 | -0.051 | -0.2398 | No |
| 65 | Slc22a5 | 17057 | -0.052 | -0.2390 | No |
| 66 | Ube2l6 | 17128 | -0.053 | -0.2394 | No |
| 67 | Erp29 | 17140 | -0.053 | -0.2376 | No |
| 68 | Aldh3a1 | 17217 | -0.055 | -0.2382 | No |
| 69 | Nbn | 17248 | -0.056 | -0.2370 | No |
| 70 | Reep6 | 17538 | -0.062 | -0.2450 | No |
| 71 | Alad | 17608 | -0.063 | -0.2450 | No |
| 72 | Dlst | 17674 | -0.064 | -0.2448 | No |
| 73 | Eci1 | 17707 | -0.065 | -0.2433 | No |
| 74 | Gabarapl1 | 17864 | -0.069 | -0.2463 | No |
| 75 | Hmgcs1 | 18258 | -0.077 | -0.2575 | No |
| 76 | Idi1 | 18334 | -0.079 | -0.2570 | No |
| 77 | Gcdh | 18342 | -0.079 | -0.2541 | No |
| 78 | Sucla2 | 18402 | -0.081 | -0.2530 | No |
| 79 | Retsat | 18522 | -0.084 | -0.2539 | No |
| 80 | Hpgd | 18855 | -0.091 | -0.2623 | No |
| 81 | Ldha | 18986 | -0.094 | -0.2633 | No |
| 82 | Serinc1 | 18990 | -0.094 | -0.2596 | No |
| 83 | Acat2 | 19149 | -0.098 | -0.2614 | No |
| 84 | Hsd17b7 | 19462 | -0.106 | -0.2685 | Yes |
| 85 | Hadhb | 19500 | -0.107 | -0.2655 | Yes |
| 86 | Idh3g | 19552 | -0.108 | -0.2630 | Yes |
| 87 | Acaa2 | 19576 | -0.109 | -0.2595 | Yes |
| 88 | Pdha1 | 19671 | -0.111 | -0.2584 | Yes |
| 89 | Car6 | 19899 | -0.116 | -0.2620 | Yes |
| 90 | Eno3 | 19918 | -0.116 | -0.2579 | Yes |
| 91 | Elovl5 | 19960 | -0.117 | -0.2547 | Yes |
| 92 | Hsp90aa1 | 20078 | -0.120 | -0.2541 | Yes |
| 93 | Aco2 | 20199 | -0.123 | -0.2536 | Yes |
| 94 | Mdh2 | 20322 | -0.126 | -0.2529 | Yes |
| 95 | Fh1 | 20407 | -0.128 | -0.2508 | Yes |
| 96 | Gstz1 | 20587 | -0.133 | -0.2520 | Yes |
| 97 | Sdhd | 20782 | -0.139 | -0.2535 | Yes |
| 98 | Cpt1a | 20848 | -0.140 | -0.2502 | Yes |
| 99 | Idh3b | 20927 | -0.142 | -0.2473 | Yes |
| 100 | Glul | 21100 | -0.147 | -0.2476 | Yes |
| 101 | Prdx6 | 21127 | -0.148 | -0.2425 | Yes |
| 102 | Ltc4s | 21133 | -0.149 | -0.2367 | Yes |
| 103 | Decr1 | 21328 | -0.155 | -0.2375 | Yes |
| 104 | Urod | 21372 | -0.156 | -0.2328 | Yes |
| 105 | Etfdh | 21408 | -0.157 | -0.2277 | Yes |
| 106 | Aldh3a2 | 21525 | -0.161 | -0.2255 | Yes |
| 107 | Hsdl2 | 21585 | -0.162 | -0.2210 | Yes |
| 108 | Aldh9a1 | 21595 | -0.163 | -0.2148 | Yes |
| 109 | Aqp7 | 21756 | -0.168 | -0.2138 | Yes |
| 110 | Sdhc | 21849 | -0.172 | -0.2102 | Yes |
| 111 | Aldh1a1 | 21948 | -0.175 | -0.2068 | Yes |
| 112 | Hmgcl | 22041 | -0.176 | -0.2030 | Yes |
| 113 | Cpox | 22046 | -0.177 | -0.1960 | Yes |
| 114 | Apex1 | 22272 | -0.185 | -0.1967 | Yes |
| 115 | Hadh | 22277 | -0.185 | -0.1894 | Yes |
| 116 | Eci2 | 22353 | -0.188 | -0.1846 | Yes |
| 117 | Ugdh | 22535 | -0.195 | -0.1833 | Yes |
| 118 | Mif | 22900 | -0.209 | -0.1881 | Yes |
| 119 | Fmo1 | 22924 | -0.210 | -0.1805 | Yes |
| 120 | Rdh11 | 23021 | -0.213 | -0.1754 | Yes |
| 121 | S100a10 | 23419 | -0.230 | -0.1805 | Yes |
| 122 | Hsd17b10 | 23504 | -0.234 | -0.1742 | Yes |
| 123 | Car2 | 23563 | -0.236 | -0.1667 | Yes |
| 124 | Hsd17b4 | 23567 | -0.236 | -0.1573 | Yes |
| 125 | Cryz | 23648 | -0.240 | -0.1505 | Yes |
| 126 | Acads | 23668 | -0.241 | -0.1415 | Yes |
| 127 | Hsd17b11 | 23737 | -0.244 | -0.1341 | Yes |
| 128 | Cbr1 | 23894 | -0.252 | -0.1296 | Yes |
| 129 | Ech1 | 24168 | -0.263 | -0.1289 | Yes |
| 130 | Mdh1 | 24442 | -0.277 | -0.1277 | Yes |
| 131 | Acot2 | 24555 | -0.282 | -0.1204 | Yes |
| 132 | Echs1 | 24566 | -0.283 | -0.1093 | Yes |
| 133 | Ostc | 24656 | -0.287 | -0.1010 | Yes |
| 134 | Acadvl | 24792 | -0.294 | -0.0940 | Yes |
| 135 | Cd36 | 24959 | -0.304 | -0.0878 | Yes |
| 136 | Gapdhs | 24992 | -0.305 | -0.0767 | Yes |
| 137 | Suclg1 | 25046 | -0.309 | -0.0661 | Yes |
| 138 | Cpt2 | 25216 | -0.319 | -0.0594 | Yes |
| 139 | Grhpr | 25438 | -0.335 | -0.0540 | Yes |
| 140 | Pts | 25558 | -0.343 | -0.0445 | Yes |
| 141 | Ppara | 25744 | -0.358 | -0.0368 | Yes |
| 142 | Psme1 | 26292 | -0.410 | -0.0401 | Yes |
| 143 | Tdo2 | 26333 | -0.414 | -0.0248 | Yes |
| 144 | Trp53inp2 | 26717 | -0.467 | -0.0199 | Yes |
| 145 | Cyp1a1 | 27066 | -0.533 | -0.0111 | Yes |
| 146 | Cidea | 27517 | -0.758 | 0.0031 | Yes |