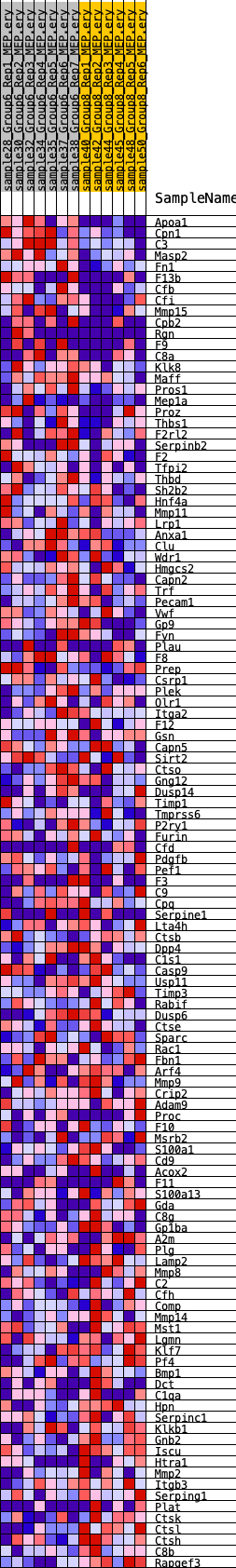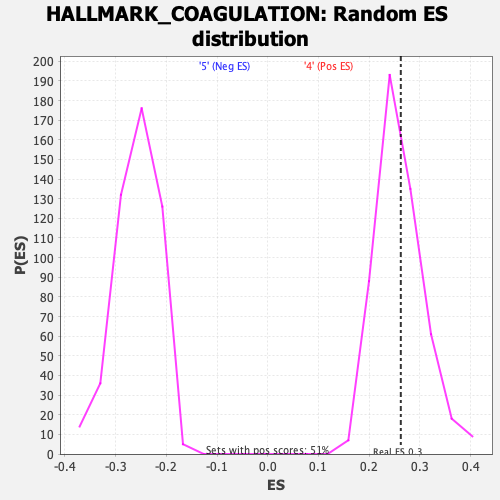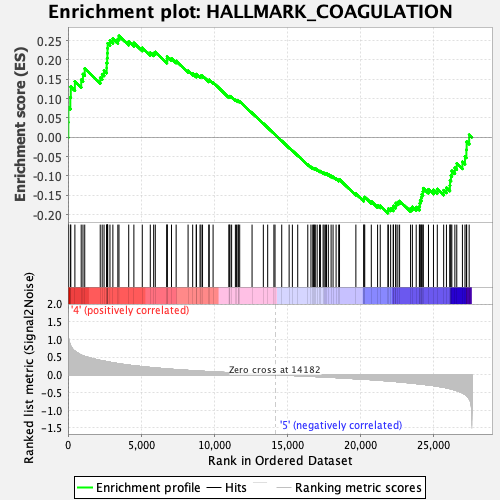
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group6_versus_Group8.MEP.ery_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.ery_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.26230982 |
| Normalized Enrichment Score (NES) | 1.0039551 |
| Nominal p-value | 0.42857143 |
| FDR q-value | 0.8102949 |
| FWER p-Value | 0.975 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Apoa1 | 21 | 1.097 | 0.0387 | Yes |
| 2 | Cpn1 | 27 | 1.077 | 0.0773 | Yes |
| 3 | C3 | 158 | 0.847 | 0.1031 | Yes |
| 4 | Masp2 | 191 | 0.803 | 0.1308 | Yes |
| 5 | Fn1 | 471 | 0.663 | 0.1445 | Yes |
| 6 | F13b | 902 | 0.553 | 0.1488 | Yes |
| 7 | Cfb | 1023 | 0.535 | 0.1637 | Yes |
| 8 | Cfi | 1144 | 0.517 | 0.1779 | Yes |
| 9 | Mmp15 | 2203 | 0.404 | 0.1539 | Yes |
| 10 | Cpb2 | 2336 | 0.393 | 0.1633 | Yes |
| 11 | Rgn | 2465 | 0.383 | 0.1724 | Yes |
| 12 | F9 | 2642 | 0.370 | 0.1793 | Yes |
| 13 | C8a | 2644 | 0.370 | 0.1926 | Yes |
| 14 | Klk8 | 2677 | 0.367 | 0.2046 | Yes |
| 15 | Maff | 2693 | 0.366 | 0.2173 | Yes |
| 16 | Pros1 | 2702 | 0.366 | 0.2302 | Yes |
| 17 | Mep1a | 2712 | 0.365 | 0.2430 | Yes |
| 18 | Proz | 2868 | 0.352 | 0.2500 | Yes |
| 19 | Thbs1 | 3067 | 0.339 | 0.2550 | Yes |
| 20 | F2rl2 | 3407 | 0.314 | 0.2540 | Yes |
| 21 | Serpinb2 | 3484 | 0.309 | 0.2623 | Yes |
| 22 | F2 | 4150 | 0.271 | 0.2479 | No |
| 23 | Tfpi2 | 4503 | 0.252 | 0.2441 | No |
| 24 | Thbd | 5077 | 0.228 | 0.2315 | No |
| 25 | Sh2b2 | 5625 | 0.204 | 0.2189 | No |
| 26 | Hnf4a | 5855 | 0.196 | 0.2177 | No |
| 27 | Mmp11 | 5966 | 0.192 | 0.2206 | No |
| 28 | Lrp1 | 6760 | 0.164 | 0.1976 | No |
| 29 | Anxa1 | 6769 | 0.164 | 0.2032 | No |
| 30 | Clu | 6777 | 0.164 | 0.2089 | No |
| 31 | Wdr1 | 7074 | 0.155 | 0.2037 | No |
| 32 | Hmgcs2 | 7392 | 0.144 | 0.1973 | No |
| 33 | Capn2 | 8208 | 0.122 | 0.1720 | No |
| 34 | Trf | 8522 | 0.113 | 0.1647 | No |
| 35 | Pecam1 | 8769 | 0.107 | 0.1596 | No |
| 36 | Vwf | 8774 | 0.107 | 0.1634 | No |
| 37 | Gp9 | 9025 | 0.101 | 0.1579 | No |
| 38 | Fyn | 9091 | 0.099 | 0.1591 | No |
| 39 | Plau | 9196 | 0.097 | 0.1588 | No |
| 40 | F8 | 9624 | 0.087 | 0.1464 | No |
| 41 | Prep | 9651 | 0.087 | 0.1486 | No |
| 42 | Csrp1 | 9917 | 0.081 | 0.1418 | No |
| 43 | Plek | 11005 | 0.059 | 0.1044 | No |
| 44 | Olr1 | 11014 | 0.059 | 0.1062 | No |
| 45 | Itga2 | 11071 | 0.058 | 0.1063 | No |
| 46 | F12 | 11184 | 0.055 | 0.1042 | No |
| 47 | Gsn | 11442 | 0.050 | 0.0966 | No |
| 48 | Capn5 | 11522 | 0.049 | 0.0955 | No |
| 49 | Sirt2 | 11630 | 0.047 | 0.0933 | No |
| 50 | Ctso | 11636 | 0.047 | 0.0948 | No |
| 51 | Gng12 | 11736 | 0.045 | 0.0928 | No |
| 52 | Dusp14 | 12581 | 0.028 | 0.0631 | No |
| 53 | Timp1 | 13352 | 0.015 | 0.0356 | No |
| 54 | Tmprss6 | 13649 | 0.009 | 0.0252 | No |
| 55 | P2ry1 | 14074 | 0.002 | 0.0098 | No |
| 56 | Furin | 14150 | 0.001 | 0.0071 | No |
| 57 | Cfd | 14609 | -0.006 | -0.0093 | No |
| 58 | Pdgfb | 15115 | -0.015 | -0.0272 | No |
| 59 | Pef1 | 15337 | -0.019 | -0.0346 | No |
| 60 | F3 | 15702 | -0.026 | -0.0469 | No |
| 61 | C9 | 16390 | -0.038 | -0.0705 | No |
| 62 | Cpq | 16607 | -0.043 | -0.0768 | No |
| 63 | Serpine1 | 16731 | -0.045 | -0.0797 | No |
| 64 | Lta4h | 16786 | -0.046 | -0.0800 | No |
| 65 | Ctsb | 16857 | -0.048 | -0.0808 | No |
| 66 | Dpp4 | 16902 | -0.048 | -0.0806 | No |
| 67 | C1s1 | 17030 | -0.051 | -0.0834 | No |
| 68 | Casp9 | 17194 | -0.055 | -0.0874 | No |
| 69 | Usp11 | 17255 | -0.056 | -0.0875 | No |
| 70 | Timp3 | 17437 | -0.059 | -0.0920 | No |
| 71 | Rabif | 17510 | -0.061 | -0.0924 | No |
| 72 | Dusp6 | 17610 | -0.063 | -0.0938 | No |
| 73 | Ctse | 17669 | -0.064 | -0.0936 | No |
| 74 | Sparc | 17795 | -0.067 | -0.0957 | No |
| 75 | Rac1 | 17984 | -0.071 | -0.1000 | No |
| 76 | Fbn1 | 18118 | -0.074 | -0.1022 | No |
| 77 | Arf4 | 18325 | -0.079 | -0.1068 | No |
| 78 | Mmp9 | 18504 | -0.083 | -0.1103 | No |
| 79 | Crip2 | 18559 | -0.085 | -0.1092 | No |
| 80 | Adam9 | 19677 | -0.111 | -0.1459 | No |
| 81 | Proc | 20214 | -0.123 | -0.1610 | No |
| 82 | F10 | 20233 | -0.123 | -0.1572 | No |
| 83 | Msrb2 | 20283 | -0.124 | -0.1545 | No |
| 84 | S100a1 | 20732 | -0.137 | -0.1659 | No |
| 85 | Cd9 | 21161 | -0.150 | -0.1760 | No |
| 86 | Acox2 | 21343 | -0.155 | -0.1770 | No |
| 87 | F11 | 21868 | -0.173 | -0.1899 | No |
| 88 | S100a13 | 21897 | -0.173 | -0.1847 | No |
| 89 | Gda | 22054 | -0.177 | -0.1840 | No |
| 90 | C8g | 22231 | -0.183 | -0.1838 | No |
| 91 | Gp1ba | 22244 | -0.184 | -0.1776 | No |
| 92 | A2m | 22400 | -0.190 | -0.1764 | No |
| 93 | Plg | 22411 | -0.190 | -0.1699 | No |
| 94 | Lamp2 | 22546 | -0.196 | -0.1678 | No |
| 95 | Mmp8 | 22666 | -0.200 | -0.1649 | No |
| 96 | C2 | 23415 | -0.230 | -0.1839 | No |
| 97 | Cfh | 23539 | -0.235 | -0.1799 | No |
| 98 | Comp | 23801 | -0.246 | -0.1805 | No |
| 99 | Mmp14 | 24018 | -0.257 | -0.1791 | No |
| 100 | Mst1 | 24045 | -0.258 | -0.1708 | No |
| 101 | Lgmn | 24074 | -0.259 | -0.1625 | No |
| 102 | Klf7 | 24147 | -0.262 | -0.1557 | No |
| 103 | Pf4 | 24184 | -0.264 | -0.1475 | No |
| 104 | Bmp1 | 24261 | -0.267 | -0.1406 | No |
| 105 | Dct | 24281 | -0.269 | -0.1316 | No |
| 106 | C1qa | 24640 | -0.286 | -0.1344 | No |
| 107 | Hpn | 24981 | -0.305 | -0.1358 | No |
| 108 | Serpinc1 | 25243 | -0.321 | -0.1337 | No |
| 109 | Klkb1 | 25678 | -0.353 | -0.1368 | No |
| 110 | Gnb2 | 25868 | -0.370 | -0.1304 | No |
| 111 | Iscu | 26104 | -0.390 | -0.1249 | No |
| 112 | Htra1 | 26115 | -0.391 | -0.1112 | No |
| 113 | Mmp2 | 26176 | -0.397 | -0.0991 | No |
| 114 | Itgb3 | 26237 | -0.403 | -0.0868 | No |
| 115 | Serping1 | 26444 | -0.428 | -0.0789 | No |
| 116 | Plat | 26574 | -0.446 | -0.0676 | No |
| 117 | Ctsk | 26960 | -0.510 | -0.0632 | No |
| 118 | Ctsl | 27135 | -0.552 | -0.0497 | No |
| 119 | Ctsh | 27230 | -0.583 | -0.0321 | No |
| 120 | C8b | 27250 | -0.593 | -0.0115 | No |
| 121 | Rapgef3 | 27421 | -0.674 | 0.0066 | No |