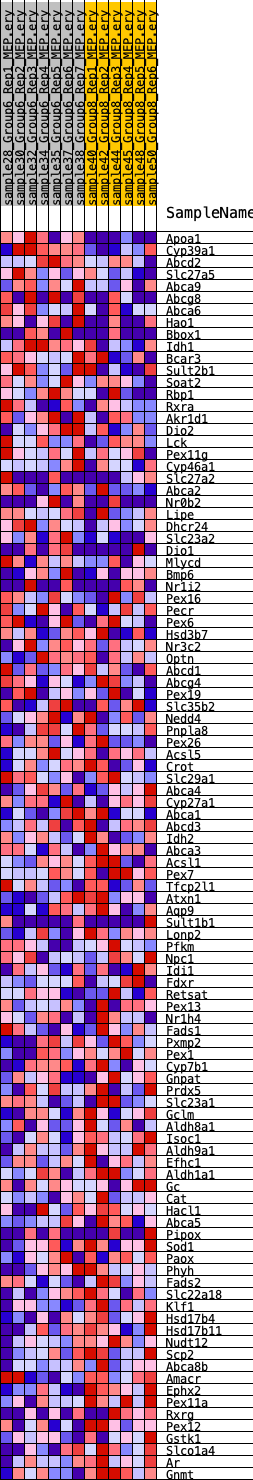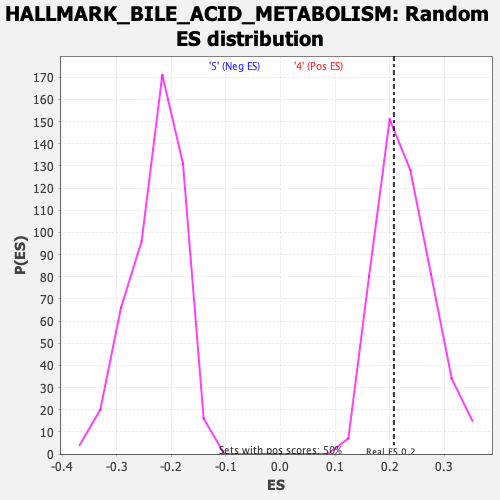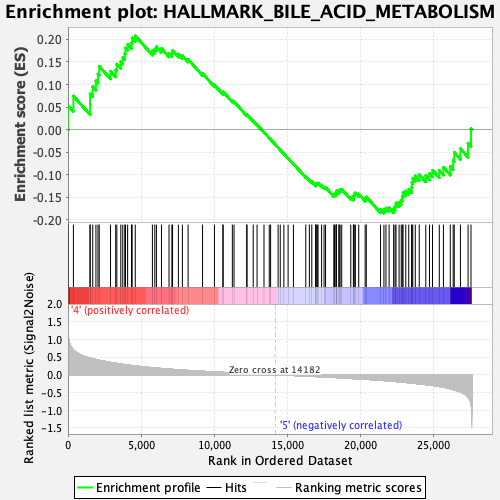
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group6_versus_Group8.MEP.ery_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | MEP.ery_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.20768616 |
| Normalized Enrichment Score (NES) | 0.91083455 |
| Nominal p-value | 0.61290324 |
| FDR q-value | 0.68346924 |
| FWER p-Value | 0.988 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Apoa1 | 21 | 1.097 | 0.0529 | Yes |
| 2 | Cyp39a1 | 369 | 0.698 | 0.0745 | Yes |
| 3 | Abcd2 | 1509 | 0.473 | 0.0562 | Yes |
| 4 | Slc27a5 | 1511 | 0.473 | 0.0793 | Yes |
| 5 | Abca9 | 1691 | 0.453 | 0.0949 | Yes |
| 6 | Abcg8 | 1909 | 0.431 | 0.1081 | Yes |
| 7 | Abca6 | 2049 | 0.417 | 0.1234 | Yes |
| 8 | Hao1 | 2134 | 0.410 | 0.1404 | Yes |
| 9 | Bbox1 | 2907 | 0.349 | 0.1294 | Yes |
| 10 | Idh1 | 3260 | 0.325 | 0.1325 | Yes |
| 11 | Bcar3 | 3338 | 0.319 | 0.1453 | Yes |
| 12 | Sult2b1 | 3609 | 0.302 | 0.1502 | Yes |
| 13 | Soat2 | 3743 | 0.293 | 0.1597 | Yes |
| 14 | Rbp1 | 3880 | 0.286 | 0.1687 | Yes |
| 15 | Rxra | 3929 | 0.284 | 0.1809 | Yes |
| 16 | Akr1d1 | 4076 | 0.275 | 0.1890 | Yes |
| 17 | Dio2 | 4340 | 0.261 | 0.1922 | Yes |
| 18 | Lck | 4384 | 0.258 | 0.2032 | Yes |
| 19 | Pex11g | 4597 | 0.248 | 0.2077 | Yes |
| 20 | Cyp46a1 | 5769 | 0.199 | 0.1748 | No |
| 21 | Slc27a2 | 5932 | 0.193 | 0.1784 | No |
| 22 | Abca2 | 6043 | 0.190 | 0.1837 | No |
| 23 | Nr0b2 | 6389 | 0.176 | 0.1797 | No |
| 24 | Lipe | 6905 | 0.159 | 0.1688 | No |
| 25 | Dhcr24 | 7101 | 0.154 | 0.1692 | No |
| 26 | Slc23a2 | 7147 | 0.152 | 0.1750 | No |
| 27 | Dio1 | 7543 | 0.140 | 0.1675 | No |
| 28 | Mlycd | 7819 | 0.131 | 0.1639 | No |
| 29 | Bmp6 | 8210 | 0.122 | 0.1557 | No |
| 30 | Nr1i2 | 9197 | 0.097 | 0.1246 | No |
| 31 | Pex16 | 10009 | 0.078 | 0.0989 | No |
| 32 | Pecr | 10584 | 0.067 | 0.0813 | No |
| 33 | Pex6 | 10605 | 0.066 | 0.0838 | No |
| 34 | Hsd3b7 | 11234 | 0.054 | 0.0636 | No |
| 35 | Nr3c2 | 11349 | 0.052 | 0.0620 | No |
| 36 | Optn | 12217 | 0.035 | 0.0322 | No |
| 37 | Abcd1 | 12235 | 0.035 | 0.0333 | No |
| 38 | Abcg4 | 12660 | 0.027 | 0.0193 | No |
| 39 | Pex19 | 12923 | 0.023 | 0.0108 | No |
| 40 | Slc35b2 | 13397 | 0.014 | -0.0057 | No |
| 41 | Nedd4 | 13757 | 0.007 | -0.0184 | No |
| 42 | Pnpla8 | 13849 | 0.006 | -0.0214 | No |
| 43 | Pex26 | 14362 | -0.001 | -0.0399 | No |
| 44 | Acsl5 | 14509 | -0.004 | -0.0451 | No |
| 45 | Crot | 14754 | -0.008 | -0.0535 | No |
| 46 | Slc29a1 | 15045 | -0.013 | -0.0634 | No |
| 47 | Abca4 | 15409 | -0.020 | -0.0756 | No |
| 48 | Cyp27a1 | 16253 | -0.036 | -0.1046 | No |
| 49 | Abca1 | 16498 | -0.041 | -0.1114 | No |
| 50 | Abcd3 | 16664 | -0.044 | -0.1153 | No |
| 51 | Idh2 | 16933 | -0.049 | -0.1226 | No |
| 52 | Abca3 | 16948 | -0.049 | -0.1207 | No |
| 53 | Acsl1 | 16960 | -0.050 | -0.1187 | No |
| 54 | Pex7 | 17039 | -0.052 | -0.1190 | No |
| 55 | Tfcp2l1 | 17091 | -0.053 | -0.1183 | No |
| 56 | Atxn1 | 17334 | -0.057 | -0.1243 | No |
| 57 | Aqp9 | 17503 | -0.061 | -0.1274 | No |
| 58 | Sult1b1 | 17606 | -0.063 | -0.1280 | No |
| 59 | Lonp2 | 18171 | -0.075 | -0.1449 | No |
| 60 | Pfkm | 18212 | -0.076 | -0.1426 | No |
| 61 | Npc1 | 18322 | -0.078 | -0.1427 | No |
| 62 | Idi1 | 18334 | -0.079 | -0.1393 | No |
| 63 | Fdxr | 18340 | -0.079 | -0.1356 | No |
| 64 | Retsat | 18522 | -0.084 | -0.1381 | No |
| 65 | Pex13 | 18528 | -0.084 | -0.1341 | No |
| 66 | Nr1h4 | 18599 | -0.086 | -0.1325 | No |
| 67 | Fads1 | 18709 | -0.087 | -0.1322 | No |
| 68 | Pxmp2 | 19328 | -0.103 | -0.1496 | No |
| 69 | Pex1 | 19508 | -0.107 | -0.1509 | No |
| 70 | Cyp7b1 | 19525 | -0.108 | -0.1462 | No |
| 71 | Gnpat | 19562 | -0.108 | -0.1423 | No |
| 72 | Prdx5 | 19643 | -0.110 | -0.1398 | No |
| 73 | Slc23a1 | 19873 | -0.116 | -0.1424 | No |
| 74 | Gclm | 20310 | -0.125 | -0.1522 | No |
| 75 | Aldh8a1 | 20390 | -0.127 | -0.1488 | No |
| 76 | Isoc1 | 21362 | -0.156 | -0.1765 | No |
| 77 | Aldh9a1 | 21595 | -0.163 | -0.1770 | No |
| 78 | Efhc1 | 21734 | -0.168 | -0.1738 | No |
| 79 | Aldh1a1 | 21948 | -0.175 | -0.1730 | No |
| 80 | Gc | 22257 | -0.184 | -0.1752 | No |
| 81 | Cat | 22355 | -0.188 | -0.1695 | No |
| 82 | Hacl1 | 22420 | -0.190 | -0.1625 | No |
| 83 | Abca5 | 22631 | -0.199 | -0.1604 | No |
| 84 | Pipox | 22779 | -0.205 | -0.1558 | No |
| 85 | Sod1 | 22849 | -0.207 | -0.1482 | No |
| 86 | Paox | 22902 | -0.209 | -0.1398 | No |
| 87 | Phyh | 23087 | -0.216 | -0.1360 | No |
| 88 | Fads2 | 23290 | -0.224 | -0.1324 | No |
| 89 | Slc22a18 | 23485 | -0.233 | -0.1280 | No |
| 90 | Klf1 | 23508 | -0.234 | -0.1174 | No |
| 91 | Hsd17b4 | 23567 | -0.236 | -0.1079 | No |
| 92 | Hsd17b11 | 23737 | -0.244 | -0.1021 | No |
| 93 | Nudt12 | 24009 | -0.256 | -0.0995 | No |
| 94 | Scp2 | 24456 | -0.278 | -0.1021 | No |
| 95 | Abca8b | 24708 | -0.290 | -0.0971 | No |
| 96 | Amacr | 24916 | -0.301 | -0.0899 | No |
| 97 | Ephx2 | 25378 | -0.330 | -0.0905 | No |
| 98 | Pex11a | 25662 | -0.351 | -0.0836 | No |
| 99 | Rxrg | 26130 | -0.392 | -0.0814 | No |
| 100 | Pex12 | 26329 | -0.414 | -0.0684 | No |
| 101 | Gstk1 | 26402 | -0.422 | -0.0503 | No |
| 102 | Slco1a4 | 26824 | -0.483 | -0.0420 | No |
| 103 | Ar | 27346 | -0.633 | -0.0300 | No |
| 104 | Gnmt | 27545 | -0.803 | 0.0021 | No |