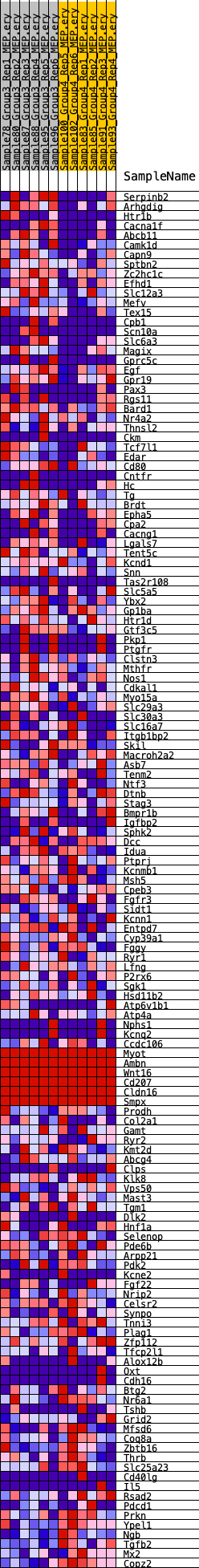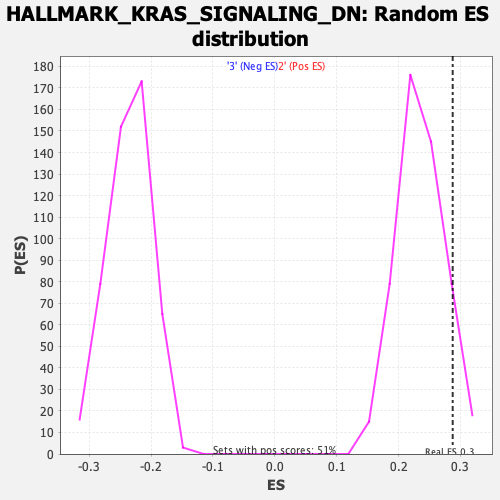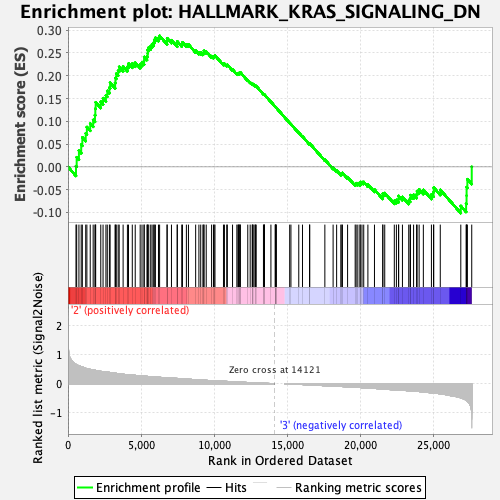
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2875413 |
| Normalized Enrichment Score (NES) | 1.2224122 |
| Nominal p-value | 0.080078125 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.95 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Serpinb2 | 546 | 0.668 | 0.0017 | Yes |
| 2 | Arhgdig | 593 | 0.653 | 0.0211 | Yes |
| 3 | Htr1b | 747 | 0.622 | 0.0357 | Yes |
| 4 | Cacna1f | 902 | 0.588 | 0.0491 | Yes |
| 5 | Abcb11 | 980 | 0.572 | 0.0647 | Yes |
| 6 | Camk1d | 1211 | 0.531 | 0.0735 | Yes |
| 7 | Capn9 | 1290 | 0.518 | 0.0875 | Yes |
| 8 | Sptbn2 | 1519 | 0.495 | 0.0952 | Yes |
| 9 | Zc2hc1c | 1720 | 0.472 | 0.1031 | Yes |
| 10 | Efhd1 | 1844 | 0.465 | 0.1137 | Yes |
| 11 | Slc12a3 | 1867 | 0.462 | 0.1278 | Yes |
| 12 | Mefv | 1892 | 0.459 | 0.1418 | Yes |
| 13 | Tex15 | 2240 | 0.427 | 0.1429 | Yes |
| 14 | Cpb1 | 2396 | 0.420 | 0.1509 | Yes |
| 15 | Scn10a | 2593 | 0.409 | 0.1570 | Yes |
| 16 | Slc6a3 | 2684 | 0.404 | 0.1667 | Yes |
| 17 | Magix | 2830 | 0.391 | 0.1741 | Yes |
| 18 | Gprc5c | 2878 | 0.387 | 0.1849 | Yes |
| 19 | Egf | 3220 | 0.364 | 0.1842 | Yes |
| 20 | Gpr19 | 3242 | 0.361 | 0.1951 | Yes |
| 21 | Pax3 | 3299 | 0.357 | 0.2046 | Yes |
| 22 | Rgs11 | 3427 | 0.346 | 0.2112 | Yes |
| 23 | Bard1 | 3498 | 0.342 | 0.2197 | Yes |
| 24 | Nr4a2 | 3767 | 0.327 | 0.2205 | Yes |
| 25 | Thnsl2 | 4061 | 0.306 | 0.2197 | Yes |
| 26 | Ckm | 4135 | 0.300 | 0.2268 | Yes |
| 27 | Tcf7l1 | 4394 | 0.296 | 0.2270 | Yes |
| 28 | Edar | 4595 | 0.287 | 0.2290 | Yes |
| 29 | Cd80 | 4934 | 0.271 | 0.2254 | Yes |
| 30 | Cntfr | 5058 | 0.265 | 0.2295 | Yes |
| 31 | Hc | 5201 | 0.265 | 0.2329 | Yes |
| 32 | Tg | 5206 | 0.264 | 0.2413 | Yes |
| 33 | Brdt | 5398 | 0.252 | 0.2425 | Yes |
| 34 | Epha5 | 5445 | 0.250 | 0.2489 | Yes |
| 35 | Cpa2 | 5449 | 0.250 | 0.2568 | Yes |
| 36 | Cacng1 | 5519 | 0.246 | 0.2623 | Yes |
| 37 | Lgals7 | 5654 | 0.241 | 0.2652 | Yes |
| 38 | Tent5c | 5748 | 0.236 | 0.2694 | Yes |
| 39 | Kcnd1 | 5859 | 0.232 | 0.2729 | Yes |
| 40 | Snn | 5898 | 0.229 | 0.2789 | Yes |
| 41 | Tas2r108 | 5974 | 0.227 | 0.2836 | Yes |
| 42 | Slc5a5 | 6185 | 0.224 | 0.2832 | Yes |
| 43 | Ybx2 | 6262 | 0.221 | 0.2875 | Yes |
| 44 | Gp1ba | 6771 | 0.208 | 0.2758 | No |
| 45 | Htr1d | 6778 | 0.208 | 0.2823 | No |
| 46 | Gtf3c5 | 7072 | 0.196 | 0.2780 | No |
| 47 | Pkp1 | 7467 | 0.181 | 0.2695 | No |
| 48 | Ptgfr | 7468 | 0.181 | 0.2753 | No |
| 49 | Clstn3 | 7777 | 0.170 | 0.2696 | No |
| 50 | Mthfr | 7823 | 0.168 | 0.2734 | No |
| 51 | Nos1 | 8092 | 0.160 | 0.2688 | No |
| 52 | Cdkal1 | 8233 | 0.154 | 0.2687 | No |
| 53 | Myo15a | 8726 | 0.139 | 0.2553 | No |
| 54 | Slc29a3 | 8948 | 0.131 | 0.2515 | No |
| 55 | Slc30a3 | 9072 | 0.127 | 0.2511 | No |
| 56 | Slc16a7 | 9222 | 0.123 | 0.2497 | No |
| 57 | Itgb1bp2 | 9282 | 0.121 | 0.2514 | No |
| 58 | Skil | 9295 | 0.121 | 0.2549 | No |
| 59 | Macroh2a2 | 9448 | 0.115 | 0.2531 | No |
| 60 | Asb7 | 9815 | 0.105 | 0.2431 | No |
| 61 | Tenm2 | 9944 | 0.101 | 0.2417 | No |
| 62 | Ntf3 | 9991 | 0.100 | 0.2433 | No |
| 63 | Dtnb | 10046 | 0.098 | 0.2445 | No |
| 64 | Stag3 | 10627 | 0.084 | 0.2261 | No |
| 65 | Bmpr1b | 10693 | 0.083 | 0.2264 | No |
| 66 | Igfbp2 | 10851 | 0.079 | 0.2232 | No |
| 67 | Sphk2 | 10891 | 0.078 | 0.2243 | No |
| 68 | Dcc | 11247 | 0.069 | 0.2136 | No |
| 69 | Idua | 11542 | 0.060 | 0.2049 | No |
| 70 | Ptprj | 11617 | 0.058 | 0.2040 | No |
| 71 | Kcnmb1 | 11657 | 0.057 | 0.2045 | No |
| 72 | Msh5 | 11695 | 0.056 | 0.2049 | No |
| 73 | Cpeb3 | 11703 | 0.056 | 0.2065 | No |
| 74 | Fgfr3 | 11772 | 0.054 | 0.2058 | No |
| 75 | Sidt1 | 11777 | 0.054 | 0.2074 | No |
| 76 | Kcnn1 | 12288 | 0.040 | 0.1901 | No |
| 77 | Entpd7 | 12452 | 0.036 | 0.1853 | No |
| 78 | Cyp39a1 | 12597 | 0.032 | 0.1811 | No |
| 79 | Fggy | 12603 | 0.032 | 0.1820 | No |
| 80 | Ryr1 | 12637 | 0.032 | 0.1818 | No |
| 81 | Lfng | 12711 | 0.030 | 0.1801 | No |
| 82 | P2rx6 | 12798 | 0.028 | 0.1779 | No |
| 83 | Sgk1 | 12812 | 0.027 | 0.1783 | No |
| 84 | Hsd11b2 | 12864 | 0.026 | 0.1773 | No |
| 85 | Atp6v1b1 | 13360 | 0.014 | 0.1597 | No |
| 86 | Atp4a | 13385 | 0.014 | 0.1593 | No |
| 87 | Nphs1 | 13427 | 0.013 | 0.1582 | No |
| 88 | Kcnq2 | 13430 | 0.013 | 0.1585 | No |
| 89 | Ccdc106 | 13870 | 0.005 | 0.1427 | No |
| 90 | Myot | 14159 | 0.000 | 0.1322 | No |
| 91 | Ambn | 14183 | 0.000 | 0.1314 | No |
| 92 | Wnt16 | 14186 | 0.000 | 0.1313 | No |
| 93 | Cd207 | 14210 | 0.000 | 0.1305 | No |
| 94 | Cldn16 | 14222 | 0.000 | 0.1301 | No |
| 95 | Smpx | 14232 | 0.000 | 0.1297 | No |
| 96 | Prodh | 15153 | -0.008 | 0.0965 | No |
| 97 | Col2a1 | 15238 | -0.010 | 0.0937 | No |
| 98 | Gamt | 15774 | -0.021 | 0.0750 | No |
| 99 | Ryr2 | 16028 | -0.028 | 0.0667 | No |
| 100 | Kmt2d | 16509 | -0.040 | 0.0505 | No |
| 101 | Abcg4 | 16525 | -0.040 | 0.0512 | No |
| 102 | Clps | 17557 | -0.067 | 0.0158 | No |
| 103 | Klk8 | 18121 | -0.080 | -0.0021 | No |
| 104 | Vps50 | 18362 | -0.086 | -0.0080 | No |
| 105 | Mast3 | 18647 | -0.094 | -0.0154 | No |
| 106 | Tgm1 | 18715 | -0.096 | -0.0147 | No |
| 107 | Dlk2 | 18759 | -0.097 | -0.0131 | No |
| 108 | Hnf1a | 19111 | -0.106 | -0.0225 | No |
| 109 | Selenop | 19629 | -0.121 | -0.0374 | No |
| 110 | Pde6b | 19695 | -0.123 | -0.0358 | No |
| 111 | Arpp21 | 19811 | -0.125 | -0.0359 | No |
| 112 | Pdk2 | 19962 | -0.131 | -0.0372 | No |
| 113 | Kcne2 | 19971 | -0.131 | -0.0332 | No |
| 114 | Fgf22 | 20094 | -0.136 | -0.0333 | No |
| 115 | Nrip2 | 20214 | -0.139 | -0.0331 | No |
| 116 | Celsr2 | 20499 | -0.148 | -0.0387 | No |
| 117 | Synpo | 20948 | -0.162 | -0.0498 | No |
| 118 | Tnni3 | 21516 | -0.182 | -0.0645 | No |
| 119 | Plag1 | 21520 | -0.182 | -0.0588 | No |
| 120 | Zfp112 | 21650 | -0.187 | -0.0574 | No |
| 121 | Tfcp2l1 | 22304 | -0.211 | -0.0744 | No |
| 122 | Alox12b | 22447 | -0.216 | -0.0726 | No |
| 123 | Oxt | 22596 | -0.221 | -0.0708 | No |
| 124 | Cdh16 | 22601 | -0.221 | -0.0638 | No |
| 125 | Btg2 | 22860 | -0.224 | -0.0660 | No |
| 126 | Nr6a1 | 23291 | -0.244 | -0.0737 | No |
| 127 | Tshb | 23384 | -0.247 | -0.0691 | No |
| 128 | Grid2 | 23410 | -0.248 | -0.0620 | No |
| 129 | Mfsd6 | 23622 | -0.258 | -0.0613 | No |
| 130 | Coq8a | 23828 | -0.262 | -0.0603 | No |
| 131 | Zbtb16 | 23861 | -0.263 | -0.0530 | No |
| 132 | Thrb | 24001 | -0.270 | -0.0493 | No |
| 133 | Slc25a23 | 24289 | -0.286 | -0.0505 | No |
| 134 | Cd40lg | 24841 | -0.314 | -0.0604 | No |
| 135 | Il5 | 24985 | -0.318 | -0.0554 | No |
| 136 | Rsad2 | 25000 | -0.319 | -0.0456 | No |
| 137 | Pdcd1 | 25446 | -0.350 | -0.0504 | No |
| 138 | Prkn | 26849 | -0.487 | -0.0857 | No |
| 139 | Ypel1 | 27213 | -0.568 | -0.0806 | No |
| 140 | Ngb | 27243 | -0.576 | -0.0630 | No |
| 141 | Tgfb2 | 27246 | -0.578 | -0.0444 | No |
| 142 | Mx2 | 27299 | -0.597 | -0.0270 | No |
| 143 | Copz2 | 27597 | -1.177 | 0.0002 | No |