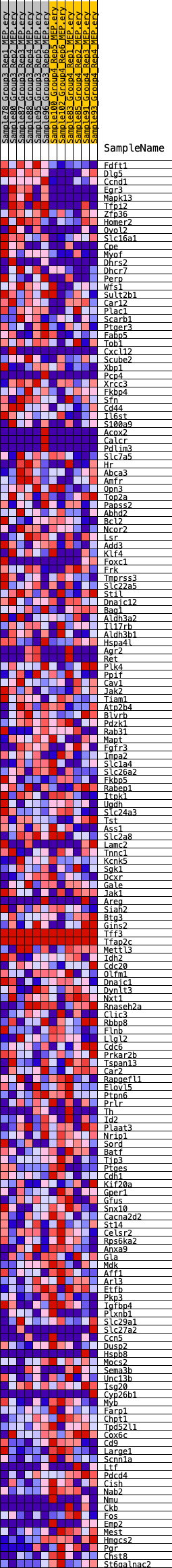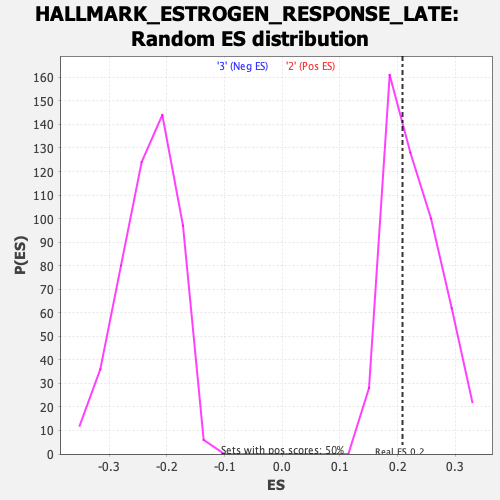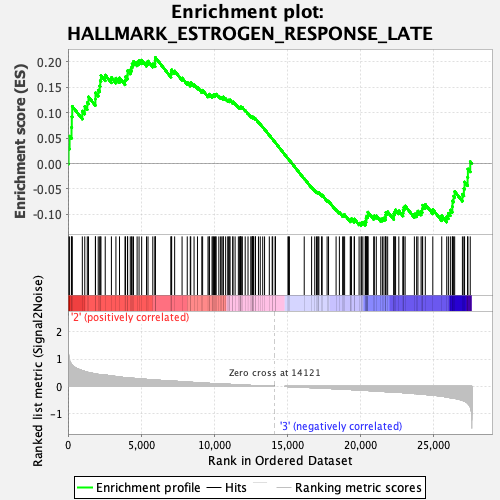
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_LATE |
| Enrichment Score (ES) | 0.20835349 |
| Normalized Enrichment Score (NES) | 0.91654664 |
| Nominal p-value | 0.59481037 |
| FDR q-value | 0.94680583 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fdft1 | 41 | 1.160 | 0.0305 | Yes |
| 2 | Dlg5 | 109 | 0.934 | 0.0538 | Yes |
| 3 | Ccnd1 | 241 | 0.797 | 0.0710 | Yes |
| 4 | Egr3 | 260 | 0.787 | 0.0920 | Yes |
| 5 | Mapk13 | 288 | 0.775 | 0.1124 | Yes |
| 6 | Tfpi2 | 979 | 0.573 | 0.1030 | Yes |
| 7 | Zfp36 | 1146 | 0.543 | 0.1120 | Yes |
| 8 | Homer2 | 1318 | 0.514 | 0.1199 | Yes |
| 9 | Ovol2 | 1399 | 0.508 | 0.1310 | Yes |
| 10 | Slc16a1 | 1872 | 0.461 | 0.1265 | Yes |
| 11 | Cpe | 1878 | 0.460 | 0.1390 | Yes |
| 12 | Myof | 2066 | 0.437 | 0.1442 | Yes |
| 13 | Dhrs2 | 2169 | 0.432 | 0.1524 | Yes |
| 14 | Dhcr7 | 2205 | 0.429 | 0.1629 | Yes |
| 15 | Perp | 2252 | 0.424 | 0.1730 | Yes |
| 16 | Wfs1 | 2545 | 0.414 | 0.1738 | Yes |
| 17 | Sult2b1 | 2967 | 0.385 | 0.1690 | Yes |
| 18 | Car12 | 3276 | 0.359 | 0.1677 | Yes |
| 19 | Plac1 | 3521 | 0.340 | 0.1681 | Yes |
| 20 | Scarb1 | 3897 | 0.318 | 0.1632 | Yes |
| 21 | Ptger3 | 3933 | 0.315 | 0.1707 | Yes |
| 22 | Fabp5 | 4072 | 0.305 | 0.1740 | Yes |
| 23 | Tob1 | 4073 | 0.305 | 0.1824 | Yes |
| 24 | Cxcl12 | 4257 | 0.300 | 0.1840 | Yes |
| 25 | Scube2 | 4346 | 0.299 | 0.1891 | Yes |
| 26 | Xbp1 | 4383 | 0.297 | 0.1959 | Yes |
| 27 | Pcp4 | 4476 | 0.292 | 0.2006 | Yes |
| 28 | Xrcc3 | 4724 | 0.280 | 0.1994 | Yes |
| 29 | Fkbp4 | 4857 | 0.275 | 0.2021 | Yes |
| 30 | Sfn | 5035 | 0.266 | 0.2030 | Yes |
| 31 | Cd44 | 5367 | 0.254 | 0.1979 | Yes |
| 32 | Il6st | 5468 | 0.249 | 0.2012 | Yes |
| 33 | S100a9 | 5797 | 0.233 | 0.1956 | Yes |
| 34 | Acox2 | 5954 | 0.227 | 0.1962 | Yes |
| 35 | Calcr | 5956 | 0.227 | 0.2024 | Yes |
| 36 | Pdlim3 | 5967 | 0.227 | 0.2084 | Yes |
| 37 | Slc7a5 | 7038 | 0.197 | 0.1748 | No |
| 38 | Hr | 7052 | 0.197 | 0.1797 | No |
| 39 | Abca3 | 7086 | 0.196 | 0.1839 | No |
| 40 | Amfr | 7290 | 0.188 | 0.1817 | No |
| 41 | Opn3 | 7797 | 0.170 | 0.1679 | No |
| 42 | Top2a | 8151 | 0.157 | 0.1594 | No |
| 43 | Papss2 | 8358 | 0.149 | 0.1560 | No |
| 44 | Abhd2 | 8389 | 0.148 | 0.1590 | No |
| 45 | Bcl2 | 8617 | 0.142 | 0.1547 | No |
| 46 | Ncor2 | 8850 | 0.134 | 0.1499 | No |
| 47 | Lsr | 9147 | 0.125 | 0.1426 | No |
| 48 | Add3 | 9202 | 0.124 | 0.1440 | No |
| 49 | Klf4 | 9570 | 0.112 | 0.1337 | No |
| 50 | Foxc1 | 9648 | 0.109 | 0.1339 | No |
| 51 | Frk | 9666 | 0.109 | 0.1363 | No |
| 52 | Tmprss3 | 9846 | 0.104 | 0.1326 | No |
| 53 | Slc22a5 | 9905 | 0.102 | 0.1334 | No |
| 54 | Stil | 9940 | 0.101 | 0.1349 | No |
| 55 | Dnajc12 | 10014 | 0.099 | 0.1350 | No |
| 56 | Bag1 | 10088 | 0.097 | 0.1350 | No |
| 57 | Aldh3a2 | 10126 | 0.096 | 0.1363 | No |
| 58 | Il17rb | 10310 | 0.092 | 0.1321 | No |
| 59 | Aldh3b1 | 10432 | 0.090 | 0.1302 | No |
| 60 | Hspa4l | 10516 | 0.087 | 0.1296 | No |
| 61 | Agr2 | 10610 | 0.085 | 0.1285 | No |
| 62 | Ret | 10611 | 0.085 | 0.1309 | No |
| 63 | Plk4 | 10776 | 0.081 | 0.1271 | No |
| 64 | Ppif | 10922 | 0.077 | 0.1239 | No |
| 65 | Cav1 | 10961 | 0.076 | 0.1246 | No |
| 66 | Jak2 | 11062 | 0.073 | 0.1230 | No |
| 67 | Tiam1 | 11064 | 0.073 | 0.1250 | No |
| 68 | Atp2b4 | 11261 | 0.068 | 0.1197 | No |
| 69 | Blvrb | 11274 | 0.068 | 0.1212 | No |
| 70 | Pdzk1 | 11422 | 0.064 | 0.1176 | No |
| 71 | Rab31 | 11644 | 0.057 | 0.1111 | No |
| 72 | Mapt | 11733 | 0.055 | 0.1094 | No |
| 73 | Fgfr3 | 11772 | 0.054 | 0.1095 | No |
| 74 | Impa2 | 11783 | 0.054 | 0.1106 | No |
| 75 | Slc1a4 | 11803 | 0.053 | 0.1114 | No |
| 76 | Slc26a2 | 11874 | 0.051 | 0.1103 | No |
| 77 | Fkbp5 | 11915 | 0.050 | 0.1102 | No |
| 78 | Rabep1 | 12112 | 0.045 | 0.1043 | No |
| 79 | Itpk1 | 12305 | 0.040 | 0.0984 | No |
| 80 | Ugdh | 12505 | 0.035 | 0.0921 | No |
| 81 | Slc24a3 | 12517 | 0.034 | 0.0926 | No |
| 82 | Tst | 12616 | 0.032 | 0.0899 | No |
| 83 | Ass1 | 12617 | 0.032 | 0.0908 | No |
| 84 | Slc2a8 | 12621 | 0.032 | 0.0916 | No |
| 85 | Lamc2 | 12666 | 0.031 | 0.0908 | No |
| 86 | Tnnc1 | 12691 | 0.030 | 0.0908 | No |
| 87 | Kcnk5 | 12808 | 0.027 | 0.0873 | No |
| 88 | Sgk1 | 12812 | 0.027 | 0.0880 | No |
| 89 | Dcxr | 13018 | 0.022 | 0.0811 | No |
| 90 | Gale | 13141 | 0.019 | 0.0772 | No |
| 91 | Jak1 | 13309 | 0.015 | 0.0715 | No |
| 92 | Areg | 13436 | 0.013 | 0.0673 | No |
| 93 | Siah2 | 13760 | 0.008 | 0.0557 | No |
| 94 | Btg3 | 13957 | 0.003 | 0.0487 | No |
| 95 | Gins2 | 13991 | 0.002 | 0.0475 | No |
| 96 | Tff3 | 14158 | 0.000 | 0.0415 | No |
| 97 | Tfap2c | 14180 | 0.000 | 0.0407 | No |
| 98 | Mettl3 | 15035 | -0.005 | 0.0097 | No |
| 99 | Idh2 | 15075 | -0.006 | 0.0085 | No |
| 100 | Cdc20 | 15134 | -0.007 | 0.0066 | No |
| 101 | Olfm1 | 16147 | -0.031 | -0.0295 | No |
| 102 | Dnajc1 | 16651 | -0.044 | -0.0466 | No |
| 103 | Dynlt3 | 16867 | -0.049 | -0.0531 | No |
| 104 | Nxt1 | 16984 | -0.052 | -0.0559 | No |
| 105 | Rnaseh2a | 17038 | -0.053 | -0.0563 | No |
| 106 | Clic3 | 17101 | -0.055 | -0.0571 | No |
| 107 | Rbbp8 | 17150 | -0.057 | -0.0573 | No |
| 108 | Flnb | 17332 | -0.061 | -0.0622 | No |
| 109 | Llgl2 | 17372 | -0.063 | -0.0619 | No |
| 110 | Cdc6 | 17711 | -0.069 | -0.0723 | No |
| 111 | Prkar2b | 17809 | -0.072 | -0.0738 | No |
| 112 | Tspan13 | 18325 | -0.085 | -0.0903 | No |
| 113 | Car2 | 18550 | -0.091 | -0.0959 | No |
| 114 | Rapgefl1 | 18772 | -0.097 | -0.1013 | No |
| 115 | Elovl5 | 18839 | -0.098 | -0.1010 | No |
| 116 | Ptpn6 | 18908 | -0.100 | -0.1007 | No |
| 117 | Prlr | 19294 | -0.112 | -0.1117 | No |
| 118 | Th | 19330 | -0.113 | -0.1098 | No |
| 119 | Id2 | 19373 | -0.114 | -0.1082 | No |
| 120 | Plaat3 | 19569 | -0.119 | -0.1120 | No |
| 121 | Nrip1 | 19574 | -0.119 | -0.1089 | No |
| 122 | Sord | 19897 | -0.128 | -0.1171 | No |
| 123 | Batf | 20030 | -0.133 | -0.1182 | No |
| 124 | Tjp3 | 20073 | -0.135 | -0.1161 | No |
| 125 | Ptges | 20155 | -0.137 | -0.1152 | No |
| 126 | Cdh1 | 20321 | -0.143 | -0.1173 | No |
| 127 | Kif20a | 20327 | -0.143 | -0.1136 | No |
| 128 | Gper1 | 20373 | -0.144 | -0.1112 | No |
| 129 | Gfus | 20376 | -0.145 | -0.1073 | No |
| 130 | Snx10 | 20395 | -0.145 | -0.1039 | No |
| 131 | Cacna2d2 | 20477 | -0.148 | -0.1028 | No |
| 132 | St14 | 20498 | -0.148 | -0.0995 | No |
| 133 | Celsr2 | 20499 | -0.148 | -0.0954 | No |
| 134 | Rps6ka2 | 20896 | -0.160 | -0.1054 | No |
| 135 | Anxa9 | 20939 | -0.162 | -0.1025 | No |
| 136 | Gla | 21073 | -0.167 | -0.1027 | No |
| 137 | Mdk | 21380 | -0.177 | -0.1090 | No |
| 138 | Aff1 | 21490 | -0.181 | -0.1080 | No |
| 139 | Arl3 | 21597 | -0.185 | -0.1068 | No |
| 140 | Etfb | 21713 | -0.189 | -0.1058 | No |
| 141 | Pkp3 | 21716 | -0.190 | -0.1006 | No |
| 142 | Igfbp4 | 21730 | -0.191 | -0.0958 | No |
| 143 | Plxnb1 | 21851 | -0.195 | -0.0948 | No |
| 144 | Slc29a1 | 22269 | -0.210 | -0.1042 | No |
| 145 | Slc27a2 | 22272 | -0.210 | -0.0985 | No |
| 146 | Ccn5 | 22335 | -0.213 | -0.0949 | No |
| 147 | Dusp2 | 22392 | -0.215 | -0.0910 | No |
| 148 | Hspb8 | 22613 | -0.221 | -0.0929 | No |
| 149 | Mocs2 | 22897 | -0.226 | -0.0970 | No |
| 150 | Sema3b | 22908 | -0.227 | -0.0911 | No |
| 151 | Unc13b | 22948 | -0.228 | -0.0863 | No |
| 152 | Isg20 | 23044 | -0.233 | -0.0833 | No |
| 153 | Cyp26b1 | 23676 | -0.260 | -0.0992 | No |
| 154 | Myb | 23831 | -0.262 | -0.0975 | No |
| 155 | Farp1 | 23918 | -0.266 | -0.0934 | No |
| 156 | Chpt1 | 24135 | -0.278 | -0.0936 | No |
| 157 | Tpd52l1 | 24216 | -0.282 | -0.0887 | No |
| 158 | Cox6c | 24242 | -0.283 | -0.0818 | No |
| 159 | Cd9 | 24423 | -0.291 | -0.0803 | No |
| 160 | Large1 | 24936 | -0.316 | -0.0903 | No |
| 161 | Scnn1a | 25546 | -0.359 | -0.1026 | No |
| 162 | Ltf | 25888 | -0.382 | -0.1045 | No |
| 163 | Pdcd4 | 26000 | -0.394 | -0.0977 | No |
| 164 | Cish | 26135 | -0.411 | -0.0912 | No |
| 165 | Nab2 | 26268 | -0.421 | -0.0845 | No |
| 166 | Nmu | 26281 | -0.422 | -0.0732 | No |
| 167 | Ckb | 26358 | -0.428 | -0.0642 | No |
| 168 | Fos | 26431 | -0.432 | -0.0549 | No |
| 169 | Emp2 | 26959 | -0.506 | -0.0602 | No |
| 170 | Mest | 27061 | -0.527 | -0.0493 | No |
| 171 | Hmgcs2 | 27107 | -0.536 | -0.0362 | No |
| 172 | Pgr | 27313 | -0.604 | -0.0270 | No |
| 173 | Chst8 | 27334 | -0.619 | -0.0107 | No |
| 174 | St6galnac2 | 27495 | -0.741 | 0.0039 | No |