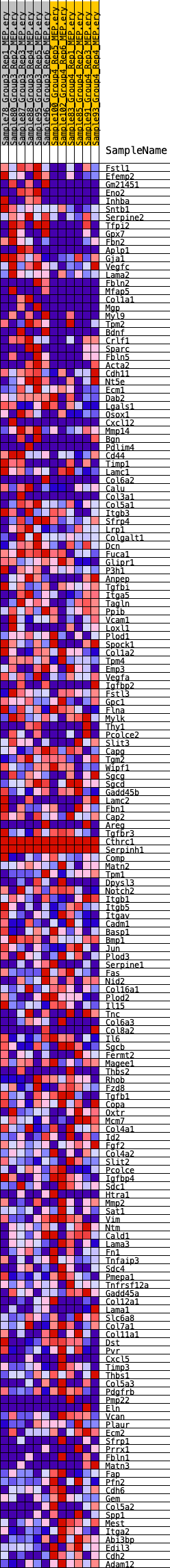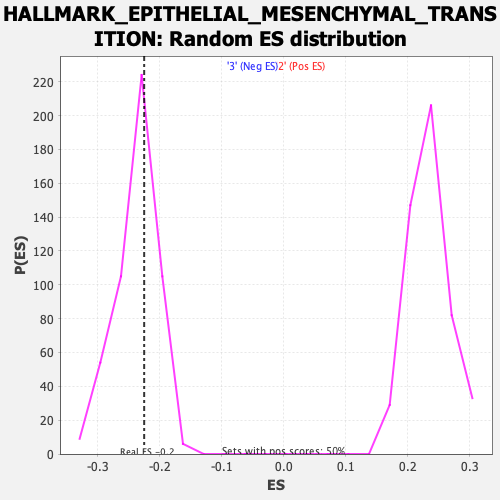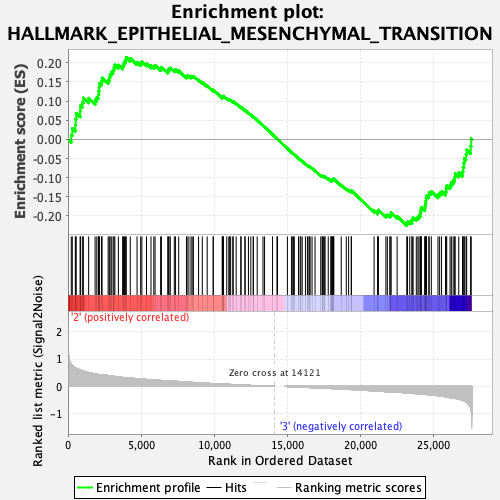
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.2250967 |
| Normalized Enrichment Score (NES) | -0.94670236 |
| Nominal p-value | 0.62823063 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fstl1 | 224 | 0.810 | 0.0117 | No |
| 2 | Efemp2 | 292 | 0.772 | 0.0283 | No |
| 3 | Gm21451 | 502 | 0.680 | 0.0374 | No |
| 4 | Eno2 | 524 | 0.675 | 0.0532 | No |
| 5 | Inhba | 558 | 0.664 | 0.0683 | No |
| 6 | Sntb1 | 834 | 0.605 | 0.0732 | No |
| 7 | Serpine2 | 843 | 0.602 | 0.0877 | No |
| 8 | Tfpi2 | 979 | 0.573 | 0.0969 | No |
| 9 | Gpx7 | 1050 | 0.563 | 0.1082 | No |
| 10 | Fbn2 | 1411 | 0.506 | 0.1075 | No |
| 11 | Aplp1 | 1852 | 0.463 | 0.1028 | No |
| 12 | Gja1 | 1969 | 0.448 | 0.1096 | No |
| 13 | Vegfc | 2076 | 0.435 | 0.1165 | No |
| 14 | Lama2 | 2097 | 0.434 | 0.1264 | No |
| 15 | Fbln2 | 2128 | 0.432 | 0.1359 | No |
| 16 | Mfap5 | 2141 | 0.432 | 0.1461 | No |
| 17 | Col1a1 | 2287 | 0.424 | 0.1512 | No |
| 18 | Mgp | 2329 | 0.421 | 0.1601 | No |
| 19 | Myl9 | 2744 | 0.398 | 0.1548 | No |
| 20 | Tpm2 | 2819 | 0.392 | 0.1617 | No |
| 21 | Bdnf | 2873 | 0.387 | 0.1693 | No |
| 22 | Crlf1 | 2958 | 0.385 | 0.1757 | No |
| 23 | Sparc | 3080 | 0.374 | 0.1805 | No |
| 24 | Fbln5 | 3140 | 0.369 | 0.1874 | No |
| 25 | Acta2 | 3203 | 0.365 | 0.1942 | No |
| 26 | Cdh11 | 3443 | 0.346 | 0.1939 | No |
| 27 | Nt5e | 3732 | 0.329 | 0.1915 | No |
| 28 | Ecm1 | 3798 | 0.325 | 0.1972 | No |
| 29 | Dab2 | 3860 | 0.321 | 0.2028 | No |
| 30 | Lgals1 | 3938 | 0.315 | 0.2078 | No |
| 31 | Qsox1 | 3984 | 0.312 | 0.2138 | No |
| 32 | Cxcl12 | 4257 | 0.300 | 0.2112 | No |
| 33 | Mmp14 | 4720 | 0.281 | 0.2013 | No |
| 34 | Bgn | 4968 | 0.269 | 0.1989 | No |
| 35 | Pdlim4 | 5050 | 0.265 | 0.2025 | No |
| 36 | Cd44 | 5367 | 0.254 | 0.1972 | No |
| 37 | Timp1 | 5673 | 0.240 | 0.1920 | No |
| 38 | Lamc1 | 5866 | 0.231 | 0.1907 | No |
| 39 | Col6a2 | 5952 | 0.227 | 0.1932 | No |
| 40 | Calu | 6332 | 0.217 | 0.1847 | No |
| 41 | Col3a1 | 6386 | 0.216 | 0.1881 | No |
| 42 | Col5a1 | 6819 | 0.206 | 0.1774 | No |
| 43 | Itgb3 | 6861 | 0.204 | 0.1809 | No |
| 44 | Sfrp4 | 6906 | 0.203 | 0.1843 | No |
| 45 | Lrp1 | 6992 | 0.199 | 0.1861 | No |
| 46 | Colgalt1 | 7265 | 0.189 | 0.1809 | No |
| 47 | Dcn | 7352 | 0.185 | 0.1823 | No |
| 48 | Fuca1 | 7567 | 0.179 | 0.1789 | No |
| 49 | Glipr1 | 8097 | 0.160 | 0.1635 | No |
| 50 | P3h1 | 8141 | 0.158 | 0.1658 | No |
| 51 | Anpep | 8263 | 0.152 | 0.1652 | No |
| 52 | Tgfbi | 8408 | 0.148 | 0.1635 | No |
| 53 | Itga5 | 8487 | 0.146 | 0.1643 | No |
| 54 | Tagln | 8571 | 0.144 | 0.1648 | No |
| 55 | Ppib | 8926 | 0.132 | 0.1551 | No |
| 56 | Vcam1 | 9180 | 0.124 | 0.1490 | No |
| 57 | Loxl1 | 9514 | 0.113 | 0.1396 | No |
| 58 | Plod1 | 9925 | 0.102 | 0.1272 | No |
| 59 | Spock1 | 9928 | 0.102 | 0.1296 | No |
| 60 | Col1a2 | 10539 | 0.087 | 0.1095 | No |
| 61 | Tpm4 | 10563 | 0.086 | 0.1108 | No |
| 62 | Emp3 | 10624 | 0.084 | 0.1106 | No |
| 63 | Vegfa | 10626 | 0.084 | 0.1127 | No |
| 64 | Igfbp2 | 10851 | 0.079 | 0.1065 | No |
| 65 | Fstl3 | 10992 | 0.075 | 0.1032 | No |
| 66 | Gpc1 | 11051 | 0.073 | 0.1029 | No |
| 67 | Flna | 11118 | 0.072 | 0.1023 | No |
| 68 | Mylk | 11266 | 0.068 | 0.0986 | No |
| 69 | Thy1 | 11292 | 0.068 | 0.0993 | No |
| 70 | Pcolce2 | 11495 | 0.062 | 0.0935 | No |
| 71 | Slit3 | 11814 | 0.053 | 0.0832 | No |
| 72 | Capg | 11829 | 0.053 | 0.0840 | No |
| 73 | Tgm2 | 12098 | 0.046 | 0.0753 | No |
| 74 | Wipf1 | 12101 | 0.046 | 0.0764 | No |
| 75 | Sgcg | 12334 | 0.039 | 0.0689 | No |
| 76 | Sgcd | 12496 | 0.035 | 0.0639 | No |
| 77 | Gadd45b | 12665 | 0.031 | 0.0585 | No |
| 78 | Lamc2 | 12666 | 0.031 | 0.0592 | No |
| 79 | Fbn1 | 12933 | 0.024 | 0.0502 | No |
| 80 | Cap2 | 13330 | 0.015 | 0.0361 | No |
| 81 | Areg | 13436 | 0.013 | 0.0326 | No |
| 82 | Tgfbr3 | 13982 | 0.003 | 0.0128 | No |
| 83 | Cthrc1 | 14283 | 0.000 | 0.0018 | No |
| 84 | Serpinh1 | 14321 | 0.000 | 0.0005 | No |
| 85 | Comp | 14996 | -0.004 | -0.0240 | No |
| 86 | Matn2 | 15274 | -0.011 | -0.0338 | No |
| 87 | Tpm1 | 15363 | -0.011 | -0.0367 | No |
| 88 | Dpysl3 | 15418 | -0.013 | -0.0384 | No |
| 89 | Notch2 | 15467 | -0.014 | -0.0398 | No |
| 90 | Itgb1 | 15757 | -0.021 | -0.0498 | No |
| 91 | Itgb5 | 15867 | -0.024 | -0.0532 | No |
| 92 | Itgav | 15917 | -0.025 | -0.0544 | No |
| 93 | Cadm1 | 16008 | -0.028 | -0.0570 | No |
| 94 | Basp1 | 16230 | -0.033 | -0.0642 | No |
| 95 | Bmp1 | 16364 | -0.037 | -0.0682 | No |
| 96 | Jun | 16413 | -0.038 | -0.0690 | No |
| 97 | Plod3 | 16522 | -0.040 | -0.0719 | No |
| 98 | Serpine1 | 16543 | -0.041 | -0.0717 | No |
| 99 | Fas | 16681 | -0.045 | -0.0755 | No |
| 100 | Nid2 | 16887 | -0.049 | -0.0818 | No |
| 101 | Col16a1 | 17276 | -0.060 | -0.0945 | No |
| 102 | Plod2 | 17370 | -0.063 | -0.0963 | No |
| 103 | Il15 | 17404 | -0.064 | -0.0960 | No |
| 104 | Tnc | 17453 | -0.065 | -0.0961 | No |
| 105 | Col6a3 | 17550 | -0.067 | -0.0980 | No |
| 106 | Col8a2 | 17564 | -0.067 | -0.0968 | No |
| 107 | Il6 | 17789 | -0.072 | -0.1032 | No |
| 108 | Sgcb | 17970 | -0.076 | -0.1079 | No |
| 109 | Fermt2 | 17995 | -0.077 | -0.1069 | No |
| 110 | Magee1 | 17999 | -0.077 | -0.1051 | No |
| 111 | Thbs2 | 18090 | -0.080 | -0.1064 | No |
| 112 | Rhob | 18096 | -0.080 | -0.1046 | No |
| 113 | Fzd8 | 18151 | -0.081 | -0.1046 | No |
| 114 | Tgfb1 | 18170 | -0.082 | -0.1033 | No |
| 115 | Copa | 18676 | -0.094 | -0.1194 | No |
| 116 | Oxtr | 19022 | -0.104 | -0.1294 | No |
| 117 | Mcm7 | 19181 | -0.108 | -0.1325 | No |
| 118 | Col4a1 | 19361 | -0.114 | -0.1362 | No |
| 119 | Id2 | 19373 | -0.114 | -0.1338 | No |
| 120 | Fgf2 | 20922 | -0.161 | -0.1863 | No |
| 121 | Col4a2 | 21167 | -0.170 | -0.1910 | No |
| 122 | Slit2 | 21171 | -0.170 | -0.1869 | No |
| 123 | Pcolce | 21219 | -0.171 | -0.1844 | No |
| 124 | Igfbp4 | 21730 | -0.191 | -0.1983 | No |
| 125 | Sdc1 | 21827 | -0.194 | -0.1971 | No |
| 126 | Htra1 | 21993 | -0.201 | -0.1981 | No |
| 127 | Mmp2 | 22072 | -0.203 | -0.1960 | No |
| 128 | Sat1 | 22074 | -0.203 | -0.1911 | No |
| 129 | Vim | 22497 | -0.218 | -0.2011 | No |
| 130 | Ntm | 23157 | -0.238 | -0.2192 | Yes |
| 131 | Cald1 | 23208 | -0.240 | -0.2152 | Yes |
| 132 | Lama3 | 23359 | -0.246 | -0.2146 | Yes |
| 133 | Fn1 | 23511 | -0.253 | -0.2139 | Yes |
| 134 | Tnfaip3 | 23521 | -0.254 | -0.2080 | Yes |
| 135 | Sdc4 | 23589 | -0.257 | -0.2041 | Yes |
| 136 | Pmepa1 | 23808 | -0.261 | -0.2056 | Yes |
| 137 | Tnfrsf12a | 23917 | -0.266 | -0.2030 | Yes |
| 138 | Gadd45a | 23991 | -0.270 | -0.1990 | Yes |
| 139 | Col12a1 | 24080 | -0.274 | -0.1955 | Yes |
| 140 | Lama1 | 24094 | -0.275 | -0.1892 | Yes |
| 141 | Slc6a8 | 24123 | -0.277 | -0.1834 | Yes |
| 142 | Col7a1 | 24153 | -0.278 | -0.1777 | Yes |
| 143 | Col11a1 | 24386 | -0.290 | -0.1790 | Yes |
| 144 | Dst | 24405 | -0.291 | -0.1725 | Yes |
| 145 | Pvr | 24442 | -0.292 | -0.1666 | Yes |
| 146 | Cxcl5 | 24459 | -0.293 | -0.1600 | Yes |
| 147 | Timp3 | 24475 | -0.294 | -0.1533 | Yes |
| 148 | Thbs1 | 24506 | -0.296 | -0.1471 | Yes |
| 149 | Col5a3 | 24667 | -0.305 | -0.1455 | Yes |
| 150 | Pdgfrb | 24683 | -0.306 | -0.1385 | Yes |
| 151 | Pmp22 | 24834 | -0.314 | -0.1362 | Yes |
| 152 | Eln | 25290 | -0.337 | -0.1445 | Yes |
| 153 | Vcan | 25403 | -0.347 | -0.1401 | Yes |
| 154 | Plaur | 25535 | -0.358 | -0.1361 | Yes |
| 155 | Ecm2 | 25804 | -0.375 | -0.1366 | Yes |
| 156 | Sfrp1 | 25844 | -0.379 | -0.1287 | Yes |
| 157 | Prrx1 | 25887 | -0.382 | -0.1208 | Yes |
| 158 | Fbln1 | 26114 | -0.408 | -0.1191 | Yes |
| 159 | Matn3 | 26204 | -0.412 | -0.1122 | Yes |
| 160 | Fap | 26328 | -0.424 | -0.1062 | Yes |
| 161 | Pfn2 | 26429 | -0.432 | -0.0992 | Yes |
| 162 | Cdh6 | 26460 | -0.437 | -0.0896 | Yes |
| 163 | Gem | 26709 | -0.465 | -0.0872 | Yes |
| 164 | Col5a2 | 26969 | -0.507 | -0.0842 | Yes |
| 165 | Spp1 | 27001 | -0.515 | -0.0726 | Yes |
| 166 | Mest | 27061 | -0.527 | -0.0618 | Yes |
| 167 | Itga2 | 27087 | -0.532 | -0.0497 | Yes |
| 168 | Abi3bp | 27206 | -0.565 | -0.0401 | Yes |
| 169 | Edil3 | 27249 | -0.579 | -0.0274 | Yes |
| 170 | Cdh2 | 27521 | -0.776 | -0.0182 | Yes |
| 171 | Adam12 | 27559 | -0.858 | 0.0016 | Yes |