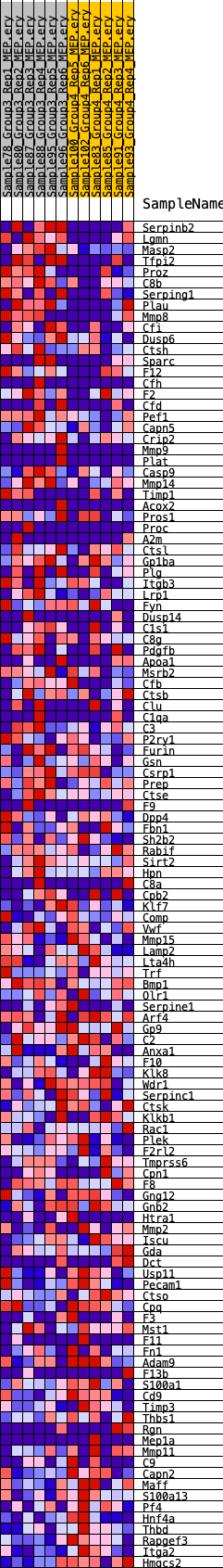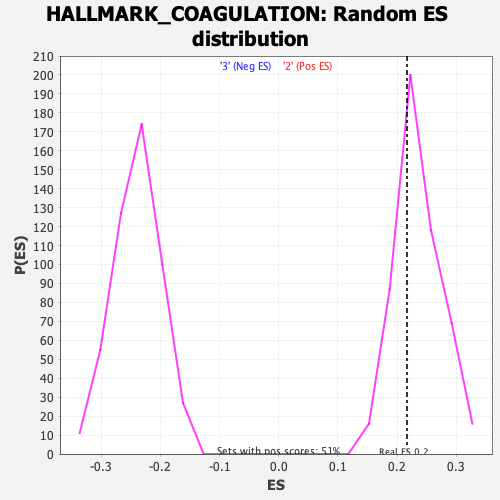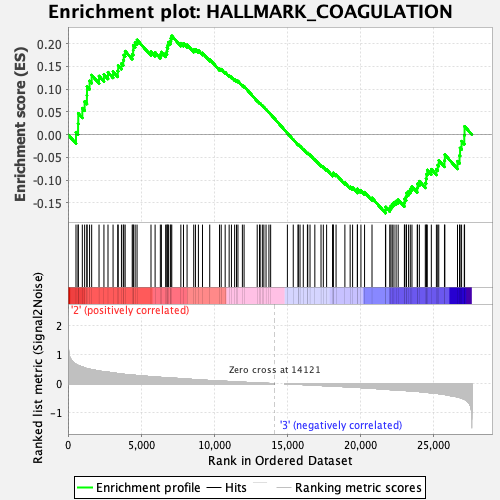
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.21676035 |
| Normalized Enrichment Score (NES) | 0.92200637 |
| Nominal p-value | 0.6936759 |
| FDR q-value | 0.9890754 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Serpinb2 | 546 | 0.668 | 0.0051 | Yes |
| 2 | Lgmn | 688 | 0.630 | 0.0236 | Yes |
| 3 | Masp2 | 701 | 0.627 | 0.0466 | Yes |
| 4 | Tfpi2 | 979 | 0.573 | 0.0579 | Yes |
| 5 | Proz | 1137 | 0.544 | 0.0726 | Yes |
| 6 | C8b | 1293 | 0.518 | 0.0863 | Yes |
| 7 | Serping1 | 1300 | 0.517 | 0.1054 | Yes |
| 8 | Plau | 1474 | 0.498 | 0.1177 | Yes |
| 9 | Mmp8 | 1618 | 0.484 | 0.1306 | Yes |
| 10 | Cfi | 2121 | 0.433 | 0.1286 | Yes |
| 11 | Dusp6 | 2456 | 0.420 | 0.1321 | Yes |
| 12 | Ctsh | 2734 | 0.399 | 0.1369 | Yes |
| 13 | Sparc | 3080 | 0.374 | 0.1384 | Yes |
| 14 | F12 | 3402 | 0.348 | 0.1397 | Yes |
| 15 | Cfh | 3430 | 0.346 | 0.1517 | Yes |
| 16 | F2 | 3653 | 0.335 | 0.1561 | Yes |
| 17 | Cfd | 3776 | 0.326 | 0.1639 | Yes |
| 18 | Pef1 | 3814 | 0.324 | 0.1746 | Yes |
| 19 | Capn5 | 3906 | 0.317 | 0.1832 | Yes |
| 20 | Crip2 | 4390 | 0.296 | 0.1767 | Yes |
| 21 | Mmp9 | 4454 | 0.292 | 0.1853 | Yes |
| 22 | Plat | 4461 | 0.292 | 0.1960 | Yes |
| 23 | Casp9 | 4588 | 0.287 | 0.2022 | Yes |
| 24 | Mmp14 | 4720 | 0.281 | 0.2079 | Yes |
| 25 | Timp1 | 5673 | 0.240 | 0.1823 | Yes |
| 26 | Acox2 | 5954 | 0.227 | 0.1806 | Yes |
| 27 | Pros1 | 6312 | 0.218 | 0.1758 | Yes |
| 28 | Proc | 6383 | 0.216 | 0.1813 | Yes |
| 29 | A2m | 6675 | 0.212 | 0.1786 | Yes |
| 30 | Ctsl | 6763 | 0.209 | 0.1833 | Yes |
| 31 | Gp1ba | 6771 | 0.208 | 0.1908 | Yes |
| 32 | Plg | 6828 | 0.206 | 0.1964 | Yes |
| 33 | Itgb3 | 6861 | 0.204 | 0.2029 | Yes |
| 34 | Lrp1 | 6992 | 0.199 | 0.2057 | Yes |
| 35 | Fyn | 7033 | 0.198 | 0.2116 | Yes |
| 36 | Dusp14 | 7093 | 0.196 | 0.2168 | Yes |
| 37 | C1s1 | 7714 | 0.172 | 0.2007 | No |
| 38 | C8g | 7894 | 0.165 | 0.2003 | No |
| 39 | Pdgfb | 8139 | 0.158 | 0.1973 | No |
| 40 | Apoa1 | 8594 | 0.142 | 0.1861 | No |
| 41 | Msrb2 | 8711 | 0.139 | 0.1871 | No |
| 42 | Cfb | 8914 | 0.132 | 0.1847 | No |
| 43 | Ctsb | 9193 | 0.124 | 0.1792 | No |
| 44 | Clu | 9688 | 0.108 | 0.1653 | No |
| 45 | C1qa | 10357 | 0.091 | 0.1444 | No |
| 46 | C3 | 10473 | 0.088 | 0.1435 | No |
| 47 | P2ry1 | 10743 | 0.081 | 0.1368 | No |
| 48 | Furin | 11015 | 0.075 | 0.1297 | No |
| 49 | Gsn | 11172 | 0.070 | 0.1267 | No |
| 50 | Csrp1 | 11391 | 0.065 | 0.1211 | No |
| 51 | Prep | 11519 | 0.061 | 0.1188 | No |
| 52 | Ctse | 11616 | 0.058 | 0.1175 | No |
| 53 | F9 | 11920 | 0.050 | 0.1083 | No |
| 54 | Dpp4 | 12036 | 0.047 | 0.1059 | No |
| 55 | Fbn1 | 12933 | 0.024 | 0.0742 | No |
| 56 | Sh2b2 | 13090 | 0.020 | 0.0693 | No |
| 57 | Rabif | 13134 | 0.019 | 0.0685 | No |
| 58 | Sirt2 | 13284 | 0.016 | 0.0636 | No |
| 59 | Hpn | 13388 | 0.013 | 0.0604 | No |
| 60 | C8a | 13537 | 0.012 | 0.0555 | No |
| 61 | Cpb2 | 13734 | 0.008 | 0.0486 | No |
| 62 | Klf7 | 13852 | 0.006 | 0.0446 | No |
| 63 | Comp | 14996 | -0.004 | 0.0032 | No |
| 64 | Vwf | 15397 | -0.012 | -0.0109 | No |
| 65 | Mmp15 | 15730 | -0.020 | -0.0223 | No |
| 66 | Lamp2 | 15736 | -0.020 | -0.0217 | No |
| 67 | Lta4h | 15860 | -0.024 | -0.0253 | No |
| 68 | Trf | 16082 | -0.029 | -0.0322 | No |
| 69 | Bmp1 | 16364 | -0.037 | -0.0411 | No |
| 70 | Olr1 | 16389 | -0.037 | -0.0406 | No |
| 71 | Serpine1 | 16543 | -0.041 | -0.0446 | No |
| 72 | Arf4 | 16872 | -0.049 | -0.0547 | No |
| 73 | Gp9 | 17295 | -0.060 | -0.0678 | No |
| 74 | C2 | 17452 | -0.065 | -0.0710 | No |
| 75 | Anxa1 | 17677 | -0.069 | -0.0766 | No |
| 76 | F10 | 18084 | -0.079 | -0.0884 | No |
| 77 | Klk8 | 18121 | -0.080 | -0.0867 | No |
| 78 | Wdr1 | 18133 | -0.080 | -0.0841 | No |
| 79 | Serpinc1 | 18323 | -0.085 | -0.0878 | No |
| 80 | Ctsk | 18926 | -0.101 | -0.1060 | No |
| 81 | Klkb1 | 19286 | -0.111 | -0.1149 | No |
| 82 | Rac1 | 19454 | -0.116 | -0.1166 | No |
| 83 | Plek | 19779 | -0.125 | -0.1237 | No |
| 84 | F2rl2 | 19783 | -0.125 | -0.1192 | No |
| 85 | Tmprss6 | 20023 | -0.133 | -0.1229 | No |
| 86 | Cpn1 | 20271 | -0.141 | -0.1266 | No |
| 87 | F8 | 20780 | -0.157 | -0.1392 | No |
| 88 | Gng12 | 21700 | -0.189 | -0.1656 | No |
| 89 | Gnb2 | 21717 | -0.190 | -0.1591 | No |
| 90 | Htra1 | 21993 | -0.201 | -0.1616 | No |
| 91 | Mmp2 | 22072 | -0.203 | -0.1568 | No |
| 92 | Iscu | 22164 | -0.206 | -0.1524 | No |
| 93 | Gda | 22281 | -0.210 | -0.1488 | No |
| 94 | Dct | 22426 | -0.216 | -0.1459 | No |
| 95 | Usp11 | 22569 | -0.221 | -0.1429 | No |
| 96 | Pecam1 | 22994 | -0.230 | -0.1497 | No |
| 97 | Ctso | 23005 | -0.231 | -0.1414 | No |
| 98 | Cpq | 23112 | -0.236 | -0.1364 | No |
| 99 | F3 | 23137 | -0.237 | -0.1284 | No |
| 100 | Mst1 | 23281 | -0.243 | -0.1245 | No |
| 101 | F11 | 23397 | -0.248 | -0.1194 | No |
| 102 | Fn1 | 23511 | -0.253 | -0.1141 | No |
| 103 | Adam9 | 23872 | -0.264 | -0.1173 | No |
| 104 | F13b | 23891 | -0.265 | -0.1081 | No |
| 105 | S100a1 | 24017 | -0.271 | -0.1025 | No |
| 106 | Cd9 | 24423 | -0.291 | -0.1063 | No |
| 107 | Timp3 | 24475 | -0.294 | -0.0972 | No |
| 108 | Thbs1 | 24506 | -0.296 | -0.0872 | No |
| 109 | Rgn | 24557 | -0.299 | -0.0779 | No |
| 110 | Mep1a | 24835 | -0.314 | -0.0762 | No |
| 111 | Mmp11 | 25181 | -0.331 | -0.0764 | No |
| 112 | C9 | 25274 | -0.336 | -0.0672 | No |
| 113 | Capn2 | 25344 | -0.342 | -0.0569 | No |
| 114 | Maff | 25733 | -0.370 | -0.0572 | No |
| 115 | S100a13 | 25759 | -0.372 | -0.0442 | No |
| 116 | Pf4 | 26623 | -0.454 | -0.0586 | No |
| 117 | Hnf4a | 26763 | -0.472 | -0.0460 | No |
| 118 | Thbd | 26799 | -0.479 | -0.0293 | No |
| 119 | Rapgef3 | 26903 | -0.495 | -0.0146 | No |
| 120 | Itga2 | 27087 | -0.532 | -0.0013 | No |
| 121 | Hmgcs2 | 27107 | -0.536 | 0.0180 | No |