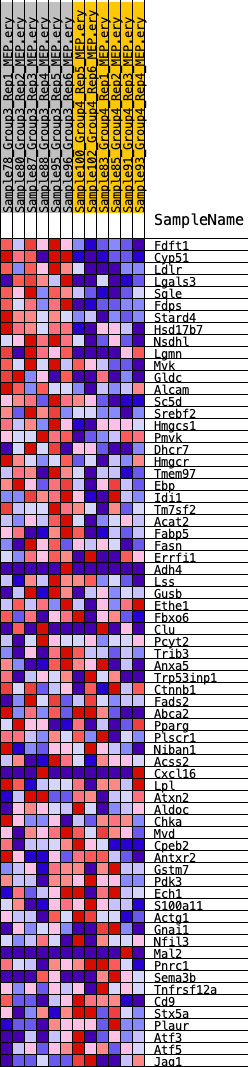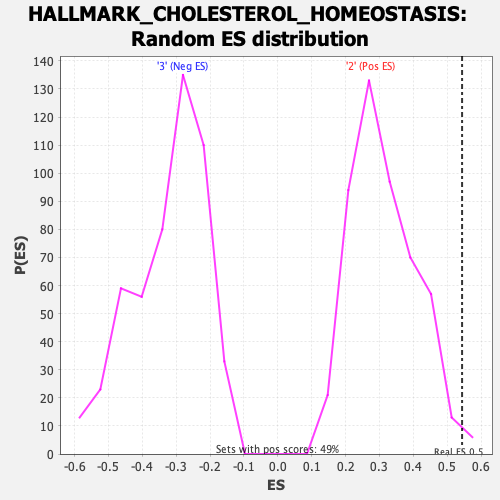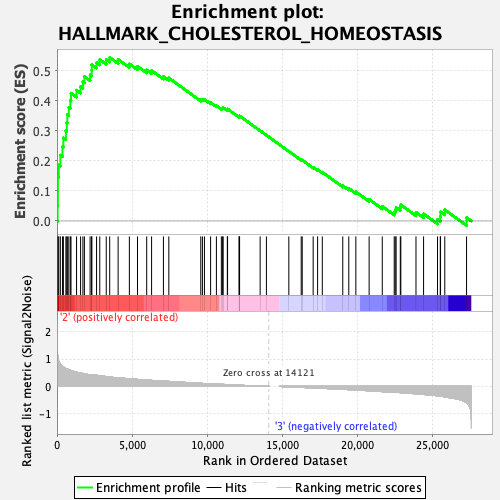
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.5438012 |
| Normalized Enrichment Score (NES) | 1.7451231 |
| Nominal p-value | 0.012219959 |
| FDR q-value | 0.21298505 |
| FWER p-Value | 0.129 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fdft1 | 41 | 1.160 | 0.0507 | Yes |
| 2 | Cyp51 | 56 | 1.087 | 0.0990 | Yes |
| 3 | Ldlr | 59 | 1.076 | 0.1474 | Yes |
| 4 | Lgals3 | 118 | 0.914 | 0.1864 | Yes |
| 5 | Sqle | 223 | 0.810 | 0.2190 | Yes |
| 6 | Fdps | 365 | 0.737 | 0.2470 | Yes |
| 7 | Stard4 | 425 | 0.709 | 0.2767 | Yes |
| 8 | Hsd17b7 | 592 | 0.653 | 0.3001 | Yes |
| 9 | Nsdhl | 653 | 0.637 | 0.3266 | Yes |
| 10 | Lgmn | 688 | 0.630 | 0.3537 | Yes |
| 11 | Mvk | 786 | 0.610 | 0.3776 | Yes |
| 12 | Gldc | 888 | 0.591 | 0.4005 | Yes |
| 13 | Alcam | 932 | 0.581 | 0.4251 | Yes |
| 14 | Sc5d | 1303 | 0.516 | 0.4349 | Yes |
| 15 | Srebf2 | 1571 | 0.489 | 0.4472 | Yes |
| 16 | Hmgcs1 | 1726 | 0.471 | 0.4628 | Yes |
| 17 | Pmvk | 1825 | 0.466 | 0.4802 | Yes |
| 18 | Dhcr7 | 2205 | 0.429 | 0.4857 | Yes |
| 19 | Hmgcr | 2310 | 0.422 | 0.5009 | Yes |
| 20 | Tmem97 | 2316 | 0.422 | 0.5197 | Yes |
| 21 | Ebp | 2636 | 0.409 | 0.5265 | Yes |
| 22 | Idi1 | 2850 | 0.389 | 0.5363 | Yes |
| 23 | Tm7sf2 | 3281 | 0.359 | 0.5368 | Yes |
| 24 | Acat2 | 3511 | 0.341 | 0.5438 | Yes |
| 25 | Fabp5 | 4072 | 0.305 | 0.5372 | No |
| 26 | Fasn | 4821 | 0.278 | 0.5225 | No |
| 27 | Errfi1 | 5363 | 0.254 | 0.5143 | No |
| 28 | Adh4 | 5976 | 0.227 | 0.5023 | No |
| 29 | Lss | 6302 | 0.219 | 0.5003 | No |
| 30 | Gusb | 7090 | 0.196 | 0.4806 | No |
| 31 | Ethe1 | 7450 | 0.182 | 0.4757 | No |
| 32 | Fbxo6 | 9580 | 0.111 | 0.4034 | No |
| 33 | Clu | 9688 | 0.108 | 0.4044 | No |
| 34 | Pcyt2 | 9823 | 0.105 | 0.4042 | No |
| 35 | Trib3 | 10235 | 0.093 | 0.3935 | No |
| 36 | Anxa5 | 10620 | 0.084 | 0.3833 | No |
| 37 | Trp53inp1 | 10949 | 0.076 | 0.3748 | No |
| 38 | Ctnnb1 | 11019 | 0.074 | 0.3757 | No |
| 39 | Fads2 | 11072 | 0.073 | 0.3771 | No |
| 40 | Abca2 | 11348 | 0.066 | 0.3700 | No |
| 41 | Pparg | 11365 | 0.065 | 0.3724 | No |
| 42 | Plscr1 | 12127 | 0.045 | 0.3468 | No |
| 43 | Niban1 | 12154 | 0.044 | 0.3478 | No |
| 44 | Acss2 | 12165 | 0.044 | 0.3494 | No |
| 45 | Cxcl16 | 13534 | 0.012 | 0.3003 | No |
| 46 | Lpl | 13952 | 0.004 | 0.2853 | No |
| 47 | Atxn2 | 15445 | -0.013 | 0.2317 | No |
| 48 | Aldoc | 16270 | -0.034 | 0.2033 | No |
| 49 | Chka | 16338 | -0.036 | 0.2025 | No |
| 50 | Mvd | 17067 | -0.054 | 0.1785 | No |
| 51 | Cpeb2 | 17363 | -0.063 | 0.1706 | No |
| 52 | Antxr2 | 17676 | -0.069 | 0.1624 | No |
| 53 | Gstm7 | 19039 | -0.104 | 0.1176 | No |
| 54 | Pdk3 | 19440 | -0.116 | 0.1083 | No |
| 55 | Ech1 | 19911 | -0.129 | 0.0970 | No |
| 56 | S100a11 | 20795 | -0.157 | 0.0720 | No |
| 57 | Actg1 | 21671 | -0.188 | 0.0487 | No |
| 58 | Gnai1 | 22467 | -0.217 | 0.0295 | No |
| 59 | Nfil3 | 22533 | -0.219 | 0.0370 | No |
| 60 | Mal2 | 22594 | -0.221 | 0.0448 | No |
| 61 | Pnrc1 | 22872 | -0.225 | 0.0449 | No |
| 62 | Sema3b | 22908 | -0.227 | 0.0538 | No |
| 63 | Tnfrsf12a | 23917 | -0.266 | 0.0291 | No |
| 64 | Cd9 | 24423 | -0.291 | 0.0239 | No |
| 65 | Stx5a | 25352 | -0.342 | 0.0056 | No |
| 66 | Plaur | 25535 | -0.358 | 0.0151 | No |
| 67 | Atf3 | 25554 | -0.360 | 0.0306 | No |
| 68 | Atf5 | 25836 | -0.378 | 0.0374 | No |
| 69 | Jag1 | 27287 | -0.594 | 0.0114 | No |