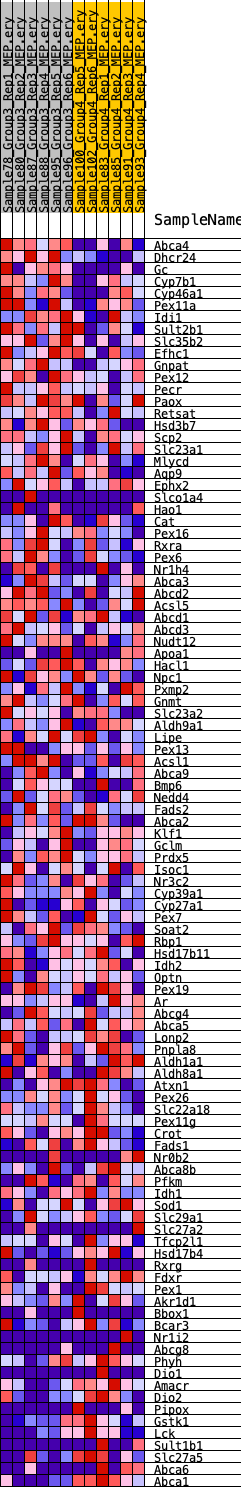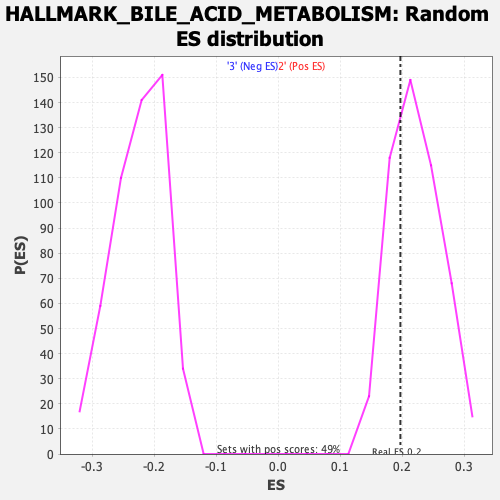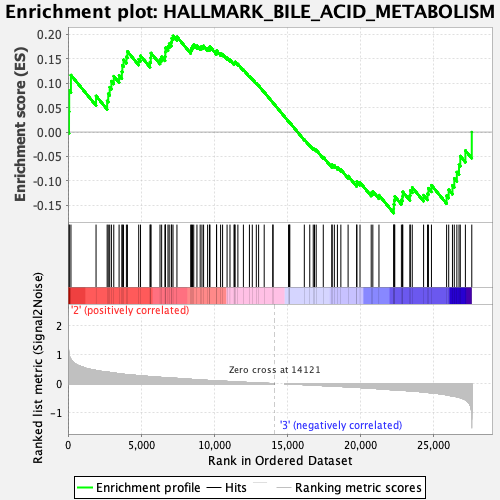
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.19671465 |
| Normalized Enrichment Score (NES) | 0.8802625 |
| Nominal p-value | 0.7008197 |
| FDR q-value | 0.9345004 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca4 | 70 | 1.028 | 0.0420 | Yes |
| 2 | Dhcr24 | 81 | 0.997 | 0.0848 | Yes |
| 3 | Gc | 201 | 0.832 | 0.1165 | Yes |
| 4 | Cyp7b1 | 1917 | 0.456 | 0.0739 | Yes |
| 5 | Cyp46a1 | 2676 | 0.405 | 0.0639 | Yes |
| 6 | Pex11a | 2750 | 0.397 | 0.0784 | Yes |
| 7 | Idi1 | 2850 | 0.389 | 0.0916 | Yes |
| 8 | Sult2b1 | 2967 | 0.385 | 0.1041 | Yes |
| 9 | Slc35b2 | 3129 | 0.370 | 0.1142 | Yes |
| 10 | Efhc1 | 3492 | 0.343 | 0.1159 | Yes |
| 11 | Gnpat | 3682 | 0.332 | 0.1234 | Yes |
| 12 | Pex12 | 3715 | 0.330 | 0.1366 | Yes |
| 13 | Pecr | 3795 | 0.326 | 0.1478 | Yes |
| 14 | Paox | 3992 | 0.311 | 0.1541 | Yes |
| 15 | Retsat | 4062 | 0.306 | 0.1649 | Yes |
| 16 | Hsd3b7 | 4833 | 0.277 | 0.1489 | Yes |
| 17 | Scp2 | 4957 | 0.270 | 0.1561 | Yes |
| 18 | Slc23a1 | 5605 | 0.243 | 0.1431 | Yes |
| 19 | Mlycd | 5656 | 0.241 | 0.1517 | Yes |
| 20 | Aqp9 | 5669 | 0.240 | 0.1616 | Yes |
| 21 | Ephx2 | 6290 | 0.219 | 0.1486 | Yes |
| 22 | Slco1a4 | 6401 | 0.216 | 0.1539 | Yes |
| 23 | Hao1 | 6631 | 0.214 | 0.1549 | Yes |
| 24 | Cat | 6654 | 0.213 | 0.1633 | Yes |
| 25 | Pex16 | 6661 | 0.213 | 0.1723 | Yes |
| 26 | Rxra | 6834 | 0.205 | 0.1749 | Yes |
| 27 | Pex6 | 6916 | 0.203 | 0.1808 | Yes |
| 28 | Nr1h4 | 7059 | 0.197 | 0.1841 | Yes |
| 29 | Abca3 | 7086 | 0.196 | 0.1917 | Yes |
| 30 | Abcd2 | 7178 | 0.193 | 0.1967 | Yes |
| 31 | Acsl5 | 7447 | 0.182 | 0.1949 | No |
| 32 | Abcd1 | 8387 | 0.149 | 0.1671 | No |
| 33 | Abcd3 | 8440 | 0.147 | 0.1716 | No |
| 34 | Nudt12 | 8511 | 0.145 | 0.1754 | No |
| 35 | Apoa1 | 8594 | 0.142 | 0.1786 | No |
| 36 | Hacl1 | 8814 | 0.135 | 0.1765 | No |
| 37 | Npc1 | 9035 | 0.128 | 0.1740 | No |
| 38 | Pxmp2 | 9156 | 0.125 | 0.1751 | No |
| 39 | Gnmt | 9268 | 0.121 | 0.1763 | No |
| 40 | Slc23a2 | 9544 | 0.113 | 0.1711 | No |
| 41 | Aldh9a1 | 9675 | 0.109 | 0.1711 | No |
| 42 | Lipe | 9702 | 0.108 | 0.1748 | No |
| 43 | Pex13 | 10150 | 0.095 | 0.1627 | No |
| 44 | Acsl1 | 10159 | 0.095 | 0.1665 | No |
| 45 | Abca9 | 10430 | 0.090 | 0.1606 | No |
| 46 | Bmp6 | 10567 | 0.086 | 0.1594 | No |
| 47 | Nedd4 | 10873 | 0.078 | 0.1517 | No |
| 48 | Fads2 | 11072 | 0.073 | 0.1476 | No |
| 49 | Abca2 | 11348 | 0.066 | 0.1405 | No |
| 50 | Klf1 | 11389 | 0.065 | 0.1418 | No |
| 51 | Gclm | 11421 | 0.064 | 0.1435 | No |
| 52 | Prdx5 | 11612 | 0.058 | 0.1391 | No |
| 53 | Isoc1 | 11989 | 0.048 | 0.1275 | No |
| 54 | Nr3c2 | 12401 | 0.038 | 0.1142 | No |
| 55 | Cyp39a1 | 12597 | 0.032 | 0.1085 | No |
| 56 | Cyp27a1 | 12872 | 0.026 | 0.0996 | No |
| 57 | Pex7 | 13029 | 0.022 | 0.0949 | No |
| 58 | Soat2 | 13411 | 0.013 | 0.0816 | No |
| 59 | Rbp1 | 13997 | 0.002 | 0.0605 | No |
| 60 | Hsd17b11 | 14017 | 0.002 | 0.0599 | No |
| 61 | Idh2 | 15075 | -0.006 | 0.0217 | No |
| 62 | Optn | 15145 | -0.008 | 0.0195 | No |
| 63 | Pex19 | 15158 | -0.008 | 0.0194 | No |
| 64 | Ar | 16153 | -0.031 | -0.0154 | No |
| 65 | Abcg4 | 16525 | -0.040 | -0.0271 | No |
| 66 | Abca5 | 16763 | -0.047 | -0.0337 | No |
| 67 | Lonp2 | 16849 | -0.049 | -0.0347 | No |
| 68 | Pnpla8 | 16965 | -0.051 | -0.0366 | No |
| 69 | Aldh1a1 | 17449 | -0.065 | -0.0514 | No |
| 70 | Aldh8a1 | 18036 | -0.078 | -0.0693 | No |
| 71 | Atxn1 | 18054 | -0.079 | -0.0666 | No |
| 72 | Pex26 | 18206 | -0.082 | -0.0685 | No |
| 73 | Slc22a18 | 18415 | -0.088 | -0.0723 | No |
| 74 | Pex11g | 18651 | -0.094 | -0.0767 | No |
| 75 | Crot | 19146 | -0.107 | -0.0901 | No |
| 76 | Fads1 | 19732 | -0.124 | -0.1060 | No |
| 77 | Nr0b2 | 19744 | -0.125 | -0.1010 | No |
| 78 | Abca8b | 19960 | -0.131 | -0.1031 | No |
| 79 | Pfkm | 20725 | -0.155 | -0.1242 | No |
| 80 | Idh1 | 20836 | -0.159 | -0.1213 | No |
| 81 | Sod1 | 21255 | -0.172 | -0.1291 | No |
| 82 | Slc29a1 | 22269 | -0.210 | -0.1568 | No |
| 83 | Slc27a2 | 22272 | -0.210 | -0.1478 | No |
| 84 | Tfcp2l1 | 22304 | -0.211 | -0.1397 | No |
| 85 | Hsd17b4 | 22336 | -0.213 | -0.1317 | No |
| 86 | Rxrg | 22789 | -0.221 | -0.1385 | No |
| 87 | Fdxr | 22868 | -0.225 | -0.1316 | No |
| 88 | Pex1 | 22883 | -0.225 | -0.1224 | No |
| 89 | Akr1d1 | 23372 | -0.247 | -0.1294 | No |
| 90 | Bbox1 | 23395 | -0.248 | -0.1195 | No |
| 91 | Bcar3 | 23531 | -0.254 | -0.1134 | No |
| 92 | Nr1i2 | 24306 | -0.287 | -0.1291 | No |
| 93 | Abcg8 | 24576 | -0.300 | -0.1259 | No |
| 94 | Phyh | 24637 | -0.303 | -0.1150 | No |
| 95 | Dio1 | 24843 | -0.314 | -0.1088 | No |
| 96 | Amacr | 25879 | -0.381 | -0.1300 | No |
| 97 | Dio2 | 26023 | -0.397 | -0.1180 | No |
| 98 | Pipox | 26279 | -0.422 | -0.1090 | No |
| 99 | Gstk1 | 26404 | -0.429 | -0.0949 | No |
| 100 | Lck | 26580 | -0.449 | -0.0818 | No |
| 101 | Sult1b1 | 26728 | -0.467 | -0.0669 | No |
| 102 | Slc27a5 | 26814 | -0.482 | -0.0491 | No |
| 103 | Abca6 | 27163 | -0.553 | -0.0378 | No |
| 104 | Abca1 | 27600 | -1.241 | 0.0001 | No |