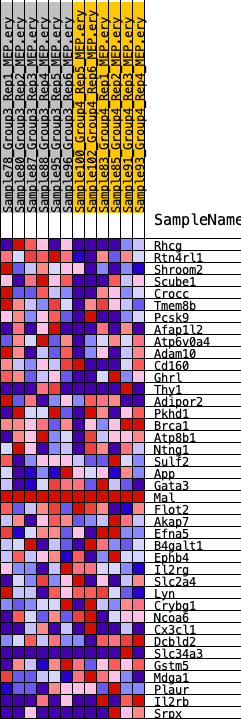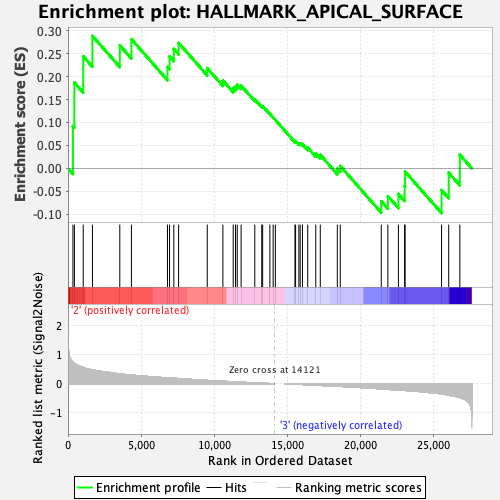
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group3_versus_Group4.MEP.ery_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_APICAL_SURFACE |
| Enrichment Score (ES) | 0.28854272 |
| Normalized Enrichment Score (NES) | 1.0546423 |
| Nominal p-value | 0.35387674 |
| FDR q-value | 0.93451715 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rhcg | 338 | 0.744 | 0.0920 | Yes |
| 2 | Rtn4rl1 | 436 | 0.703 | 0.1870 | Yes |
| 3 | Shroom2 | 1041 | 0.565 | 0.2443 | Yes |
| 4 | Scube1 | 1670 | 0.479 | 0.2885 | Yes |
| 5 | Crocc | 3541 | 0.338 | 0.2681 | No |
| 6 | Tmem8b | 4334 | 0.299 | 0.2813 | No |
| 7 | Pcsk9 | 6799 | 0.207 | 0.2209 | No |
| 8 | Afap1l2 | 6942 | 0.201 | 0.2439 | No |
| 9 | Atp6v0a4 | 7233 | 0.190 | 0.2601 | No |
| 10 | Adam10 | 7560 | 0.179 | 0.2733 | No |
| 11 | Cd160 | 9518 | 0.113 | 0.2182 | No |
| 12 | Ghrl | 10586 | 0.085 | 0.1914 | No |
| 13 | Thy1 | 11292 | 0.068 | 0.1753 | No |
| 14 | Adipor2 | 11450 | 0.063 | 0.1784 | No |
| 15 | Pkhd1 | 11568 | 0.060 | 0.1825 | No |
| 16 | Brca1 | 11833 | 0.053 | 0.1803 | No |
| 17 | Atp8b1 | 12768 | 0.028 | 0.1504 | No |
| 18 | Ntng1 | 13242 | 0.017 | 0.1357 | No |
| 19 | Sulf2 | 13305 | 0.016 | 0.1356 | No |
| 20 | App | 13799 | 0.007 | 0.1186 | No |
| 21 | Gata3 | 14022 | 0.002 | 0.1108 | No |
| 22 | Mal | 14172 | 0.000 | 0.1054 | No |
| 23 | Flot2 | 15512 | -0.015 | 0.0589 | No |
| 24 | Akap7 | 15557 | -0.016 | 0.0595 | No |
| 25 | Efna5 | 15780 | -0.022 | 0.0545 | No |
| 26 | B4galt1 | 15870 | -0.024 | 0.0546 | No |
| 27 | Ephb4 | 16025 | -0.028 | 0.0530 | No |
| 28 | Il2rg | 16387 | -0.037 | 0.0451 | No |
| 29 | Slc2a4 | 16936 | -0.051 | 0.0323 | No |
| 30 | Lyn | 17244 | -0.059 | 0.0295 | No |
| 31 | Crybg1 | 18409 | -0.087 | -0.0005 | No |
| 32 | Ncoa6 | 18610 | -0.092 | 0.0052 | No |
| 33 | Cx3cl1 | 21412 | -0.178 | -0.0715 | No |
| 34 | Dcbld2 | 21865 | -0.196 | -0.0605 | No |
| 35 | Slc34a3 | 22585 | -0.221 | -0.0556 | No |
| 36 | Gstm5 | 23009 | -0.231 | -0.0386 | No |
| 37 | Mdga1 | 23041 | -0.233 | -0.0071 | No |
| 38 | Plaur | 25535 | -0.358 | -0.0474 | No |
| 39 | Il2rb | 26024 | -0.397 | -0.0094 | No |
| 40 | Srpx | 26783 | -0.476 | 0.0297 | No |