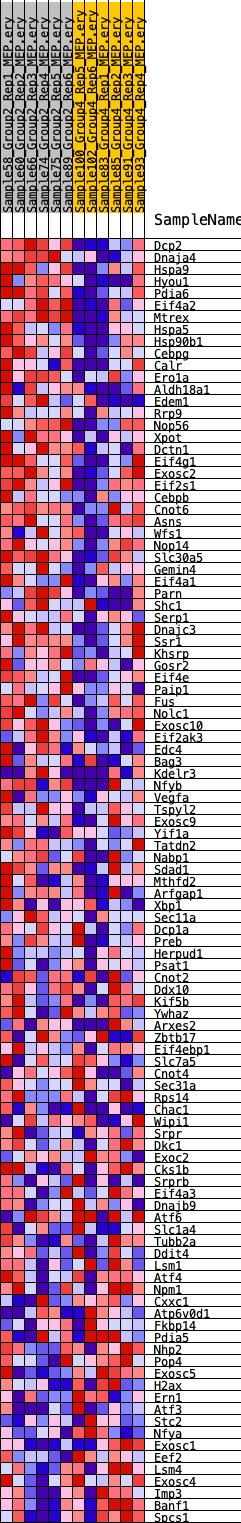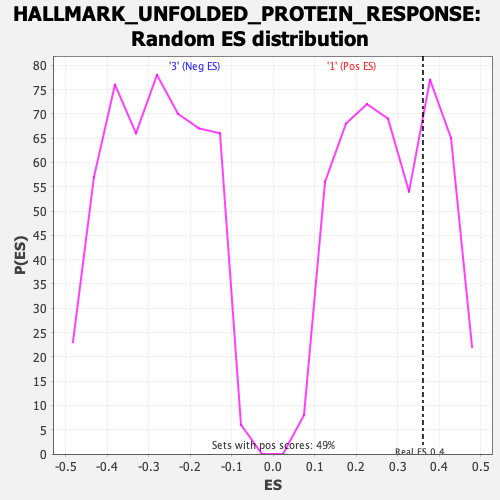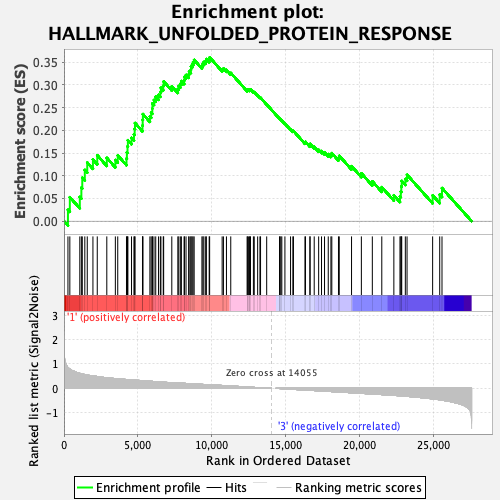
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.3606694 |
| Normalized Enrichment Score (NES) | 1.2573439 |
| Nominal p-value | 0.29735234 |
| FDR q-value | 0.5921325 |
| FWER p-Value | 0.856 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcp2 | 267 | 0.848 | 0.0251 | Yes |
| 2 | Dnaja4 | 395 | 0.788 | 0.0528 | Yes |
| 3 | Hspa9 | 1070 | 0.613 | 0.0534 | Yes |
| 4 | Hyou1 | 1176 | 0.595 | 0.0740 | Yes |
| 5 | Pdia6 | 1238 | 0.586 | 0.0958 | Yes |
| 6 | Eif4a2 | 1409 | 0.565 | 0.1128 | Yes |
| 7 | Mtrex | 1575 | 0.545 | 0.1292 | Yes |
| 8 | Hspa5 | 1959 | 0.504 | 0.1359 | Yes |
| 9 | Hsp90b1 | 2252 | 0.482 | 0.1450 | Yes |
| 10 | Cebpg | 2898 | 0.434 | 0.1394 | Yes |
| 11 | Calr | 3477 | 0.398 | 0.1347 | Yes |
| 12 | Ero1a | 3639 | 0.388 | 0.1447 | Yes |
| 13 | Aldh18a1 | 4231 | 0.359 | 0.1380 | Yes |
| 14 | Edem1 | 4258 | 0.357 | 0.1517 | Yes |
| 15 | Rrp9 | 4301 | 0.356 | 0.1647 | Yes |
| 16 | Nop56 | 4326 | 0.353 | 0.1784 | Yes |
| 17 | Xpot | 4572 | 0.342 | 0.1835 | Yes |
| 18 | Dctn1 | 4737 | 0.336 | 0.1913 | Yes |
| 19 | Eif4g1 | 4785 | 0.334 | 0.2033 | Yes |
| 20 | Exosc2 | 4807 | 0.333 | 0.2162 | Yes |
| 21 | Eif2s1 | 5315 | 0.318 | 0.2108 | Yes |
| 22 | Cebpb | 5328 | 0.317 | 0.2233 | Yes |
| 23 | Cnot6 | 5342 | 0.317 | 0.2359 | Yes |
| 24 | Asns | 5817 | 0.290 | 0.2305 | Yes |
| 25 | Wfs1 | 5901 | 0.287 | 0.2393 | Yes |
| 26 | Nop14 | 5980 | 0.283 | 0.2480 | Yes |
| 27 | Slc30a5 | 5986 | 0.283 | 0.2595 | Yes |
| 28 | Gemin4 | 6093 | 0.278 | 0.2670 | Yes |
| 29 | Eif4a1 | 6211 | 0.276 | 0.2741 | Yes |
| 30 | Parn | 6402 | 0.268 | 0.2782 | Yes |
| 31 | Shc1 | 6524 | 0.262 | 0.2845 | Yes |
| 32 | Serp1 | 6562 | 0.259 | 0.2938 | Yes |
| 33 | Dnajc3 | 6721 | 0.252 | 0.2984 | Yes |
| 34 | Ssr1 | 6748 | 0.250 | 0.3077 | Yes |
| 35 | Khsrp | 7300 | 0.231 | 0.2971 | Yes |
| 36 | Gosr2 | 7709 | 0.218 | 0.2912 | Yes |
| 37 | Eif4e | 7772 | 0.215 | 0.2978 | Yes |
| 38 | Paip1 | 7878 | 0.211 | 0.3026 | Yes |
| 39 | Fus | 7949 | 0.208 | 0.3086 | Yes |
| 40 | Nolc1 | 8135 | 0.203 | 0.3102 | Yes |
| 41 | Exosc10 | 8154 | 0.202 | 0.3178 | Yes |
| 42 | Eif2ak3 | 8263 | 0.198 | 0.3220 | Yes |
| 43 | Edc4 | 8441 | 0.191 | 0.3234 | Yes |
| 44 | Bag3 | 8473 | 0.190 | 0.3301 | Yes |
| 45 | Kdelr3 | 8591 | 0.185 | 0.3334 | Yes |
| 46 | Nfyb | 8600 | 0.185 | 0.3407 | Yes |
| 47 | Vegfa | 8678 | 0.182 | 0.3454 | Yes |
| 48 | Tspyl2 | 8742 | 0.179 | 0.3504 | Yes |
| 49 | Exosc9 | 8813 | 0.176 | 0.3551 | Yes |
| 50 | Yif1a | 9340 | 0.161 | 0.3426 | Yes |
| 51 | Tatdn2 | 9406 | 0.159 | 0.3467 | Yes |
| 52 | Nabp1 | 9472 | 0.156 | 0.3508 | Yes |
| 53 | Sdad1 | 9594 | 0.151 | 0.3526 | Yes |
| 54 | Mthfd2 | 9640 | 0.150 | 0.3571 | Yes |
| 55 | Arfgap1 | 9832 | 0.142 | 0.3560 | Yes |
| 56 | Xbp1 | 9862 | 0.141 | 0.3607 | Yes |
| 57 | Sec11a | 10709 | 0.117 | 0.3347 | No |
| 58 | Dcp1a | 10796 | 0.113 | 0.3362 | No |
| 59 | Preb | 11000 | 0.106 | 0.3332 | No |
| 60 | Herpud1 | 11292 | 0.095 | 0.3265 | No |
| 61 | Psat1 | 12410 | 0.054 | 0.2881 | No |
| 62 | Cnot2 | 12411 | 0.054 | 0.2903 | No |
| 63 | Ddx10 | 12487 | 0.051 | 0.2896 | No |
| 64 | Kif5b | 12568 | 0.048 | 0.2887 | No |
| 65 | Ywhaz | 12588 | 0.047 | 0.2899 | No |
| 66 | Arxes2 | 12629 | 0.045 | 0.2904 | No |
| 67 | Zbtb17 | 12825 | 0.038 | 0.2848 | No |
| 68 | Eif4ebp1 | 12881 | 0.036 | 0.2843 | No |
| 69 | Slc7a5 | 13113 | 0.028 | 0.2771 | No |
| 70 | Cnot4 | 13268 | 0.023 | 0.2724 | No |
| 71 | Sec31a | 13309 | 0.021 | 0.2718 | No |
| 72 | Rps14 | 13725 | 0.009 | 0.2571 | No |
| 73 | Chac1 | 14594 | -0.007 | 0.2258 | No |
| 74 | Wipi1 | 14620 | -0.008 | 0.2253 | No |
| 75 | Srpr | 14683 | -0.010 | 0.2234 | No |
| 76 | Dkc1 | 14749 | -0.013 | 0.2216 | No |
| 77 | Exoc2 | 14959 | -0.021 | 0.2149 | No |
| 78 | Cks1b | 15341 | -0.034 | 0.2024 | No |
| 79 | Srprb | 15499 | -0.040 | 0.1984 | No |
| 80 | Eif4a3 | 15537 | -0.042 | 0.1987 | No |
| 81 | Dnajb9 | 16317 | -0.069 | 0.1732 | No |
| 82 | Atf6 | 16351 | -0.069 | 0.1748 | No |
| 83 | Slc1a4 | 16653 | -0.080 | 0.1672 | No |
| 84 | Tubb2a | 16661 | -0.081 | 0.1702 | No |
| 85 | Ddit4 | 16943 | -0.090 | 0.1637 | No |
| 86 | Lsm1 | 17244 | -0.102 | 0.1570 | No |
| 87 | Atf4 | 17438 | -0.109 | 0.1544 | No |
| 88 | Npm1 | 17642 | -0.116 | 0.1518 | No |
| 89 | Cxxc1 | 17894 | -0.124 | 0.1478 | No |
| 90 | Atp6v0d1 | 18074 | -0.130 | 0.1466 | No |
| 91 | Fkbp14 | 18135 | -0.133 | 0.1499 | No |
| 92 | Pdia5 | 18586 | -0.150 | 0.1396 | No |
| 93 | Nhp2 | 18638 | -0.151 | 0.1440 | No |
| 94 | Pop4 | 19471 | -0.181 | 0.1211 | No |
| 95 | Exosc5 | 20136 | -0.205 | 0.1054 | No |
| 96 | H2ax | 20878 | -0.227 | 0.0878 | No |
| 97 | Ern1 | 21519 | -0.256 | 0.0750 | No |
| 98 | Atf3 | 22336 | -0.288 | 0.0571 | No |
| 99 | Stc2 | 22750 | -0.306 | 0.0546 | No |
| 100 | Nfya | 22808 | -0.308 | 0.0652 | No |
| 101 | Exosc1 | 22841 | -0.310 | 0.0768 | No |
| 102 | Eef2 | 22869 | -0.312 | 0.0885 | No |
| 103 | Lsm4 | 23113 | -0.322 | 0.0929 | No |
| 104 | Exosc4 | 23229 | -0.327 | 0.1022 | No |
| 105 | Imp3 | 24962 | -0.430 | 0.0568 | No |
| 106 | Banf1 | 25441 | -0.469 | 0.0587 | No |
| 107 | Spcs1 | 25596 | -0.484 | 0.0730 | No |