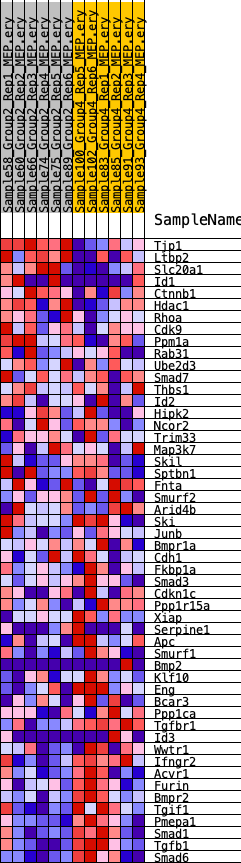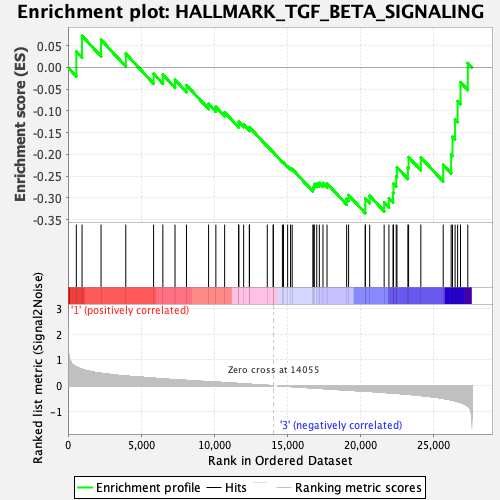
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | -0.33514696 |
| Normalized Enrichment Score (NES) | -1.1639569 |
| Nominal p-value | 0.32894737 |
| FDR q-value | 0.7044097 |
| FWER p-Value | 0.947 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tjp1 | 569 | 0.724 | 0.0365 | No |
| 2 | Ltbp2 | 960 | 0.635 | 0.0725 | No |
| 3 | Slc20a1 | 2259 | 0.482 | 0.0635 | No |
| 4 | Id1 | 3949 | 0.373 | 0.0316 | No |
| 5 | Ctnnb1 | 5850 | 0.289 | -0.0145 | No |
| 6 | Hdac1 | 6483 | 0.264 | -0.0166 | No |
| 7 | Rhoa | 7315 | 0.230 | -0.0286 | No |
| 8 | Cdk9 | 8098 | 0.205 | -0.0408 | No |
| 9 | Ppm1a | 9609 | 0.151 | -0.0837 | No |
| 10 | Rab31 | 10107 | 0.139 | -0.0908 | No |
| 11 | Ube2d3 | 10706 | 0.117 | -0.1032 | No |
| 12 | Smad7 | 11668 | 0.082 | -0.1316 | No |
| 13 | Thbs1 | 11671 | 0.082 | -0.1252 | No |
| 14 | Id2 | 12006 | 0.070 | -0.1318 | No |
| 15 | Hipk2 | 12397 | 0.054 | -0.1417 | No |
| 16 | Ncor2 | 12401 | 0.054 | -0.1375 | No |
| 17 | Trim33 | 13621 | 0.012 | -0.1808 | No |
| 18 | Map3k7 | 14025 | 0.001 | -0.1953 | No |
| 19 | Skil | 14033 | 0.001 | -0.1955 | No |
| 20 | Sptbn1 | 14655 | -0.009 | -0.2173 | No |
| 21 | Fnta | 14730 | -0.012 | -0.2190 | No |
| 22 | Smurf2 | 15013 | -0.023 | -0.2275 | No |
| 23 | Arid4b | 15212 | -0.029 | -0.2324 | No |
| 24 | Ski | 15324 | -0.034 | -0.2338 | No |
| 25 | Junb | 16737 | -0.083 | -0.2784 | No |
| 26 | Bmpr1a | 16795 | -0.086 | -0.2737 | No |
| 27 | Cdh1 | 16834 | -0.087 | -0.2682 | No |
| 28 | Fkbp1a | 17002 | -0.092 | -0.2670 | No |
| 29 | Smad3 | 17180 | -0.099 | -0.2656 | No |
| 30 | Cdkn1c | 17428 | -0.108 | -0.2660 | No |
| 31 | Ppp1r15a | 17705 | -0.117 | -0.2668 | No |
| 32 | Xiap | 19046 | -0.165 | -0.3024 | No |
| 33 | Serpine1 | 19179 | -0.170 | -0.2938 | No |
| 34 | Apc | 20320 | -0.211 | -0.3184 | Yes |
| 35 | Smurf1 | 20321 | -0.212 | -0.3017 | Yes |
| 36 | Bmp2 | 20622 | -0.221 | -0.2952 | Yes |
| 37 | Klf10 | 21610 | -0.260 | -0.3105 | Yes |
| 38 | Eng | 21936 | -0.271 | -0.3009 | Yes |
| 39 | Bcar3 | 22217 | -0.283 | -0.2887 | Yes |
| 40 | Ppp1ca | 22248 | -0.285 | -0.2673 | Yes |
| 41 | Tgfbr1 | 22433 | -0.292 | -0.2509 | Yes |
| 42 | Id3 | 22485 | -0.294 | -0.2295 | Yes |
| 43 | Wwtr1 | 23232 | -0.328 | -0.2307 | Yes |
| 44 | Ifngr2 | 23280 | -0.331 | -0.2063 | Yes |
| 45 | Acvr1 | 24117 | -0.377 | -0.2069 | Yes |
| 46 | Furin | 25646 | -0.488 | -0.2238 | Yes |
| 47 | Bmpr2 | 26190 | -0.547 | -0.2004 | Yes |
| 48 | Tgif1 | 26281 | -0.558 | -0.1596 | Yes |
| 49 | Pmepa1 | 26459 | -0.586 | -0.1197 | Yes |
| 50 | Smad1 | 26631 | -0.611 | -0.0776 | Yes |
| 51 | Tgfb1 | 26829 | -0.642 | -0.0341 | Yes |
| 52 | Smad6 | 27327 | -0.787 | 0.0100 | Yes |