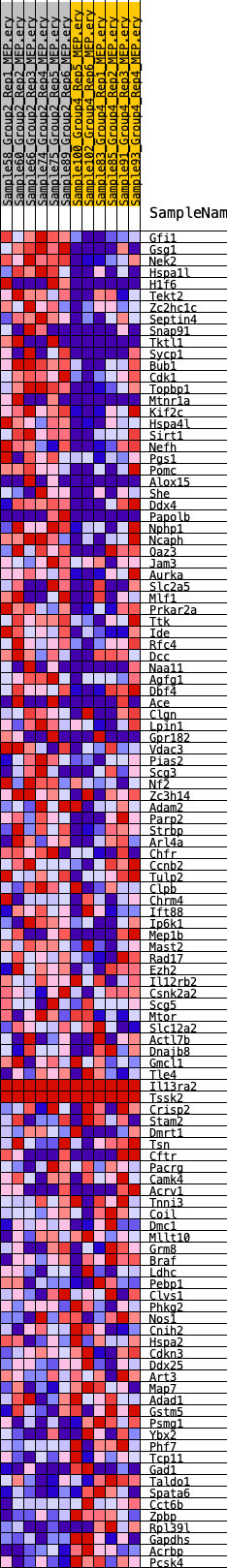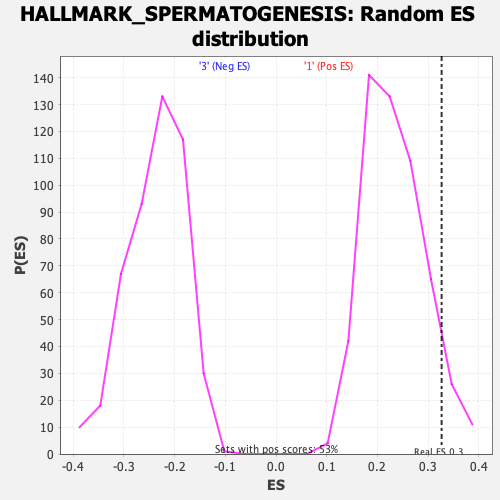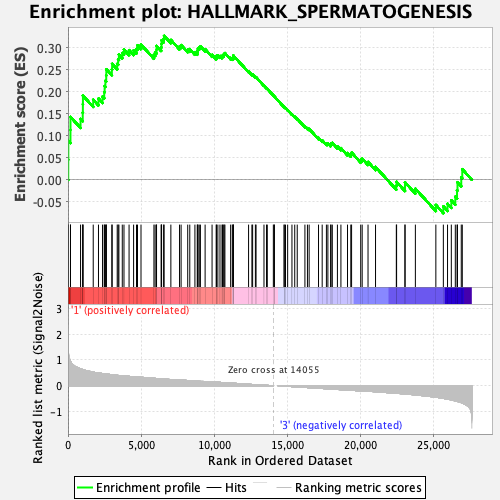
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.326838 |
| Normalized Enrichment Score (NES) | 1.3909075 |
| Nominal p-value | 0.06779661 |
| FDR q-value | 0.4213598 |
| FWER p-Value | 0.678 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gfi1 | 17 | 1.473 | 0.0455 | Yes |
| 2 | Gsg1 | 22 | 1.378 | 0.0886 | Yes |
| 3 | Nek2 | 168 | 0.948 | 0.1130 | Yes |
| 4 | Hspa1l | 171 | 0.946 | 0.1425 | Yes |
| 5 | H1f6 | 858 | 0.650 | 0.1380 | Yes |
| 6 | Tekt2 | 1007 | 0.627 | 0.1522 | Yes |
| 7 | Zc2hc1c | 1016 | 0.625 | 0.1715 | Yes |
| 8 | Septin4 | 1021 | 0.624 | 0.1909 | Yes |
| 9 | Snap91 | 1724 | 0.527 | 0.1818 | Yes |
| 10 | Tktl1 | 2083 | 0.495 | 0.1843 | Yes |
| 11 | Sycp1 | 2361 | 0.471 | 0.1890 | Yes |
| 12 | Bub1 | 2476 | 0.461 | 0.1993 | Yes |
| 13 | Cdk1 | 2504 | 0.458 | 0.2127 | Yes |
| 14 | Topbp1 | 2556 | 0.453 | 0.2250 | Yes |
| 15 | Mtnr1a | 2613 | 0.448 | 0.2370 | Yes |
| 16 | Kif2c | 2620 | 0.448 | 0.2508 | Yes |
| 17 | Hspa4l | 2998 | 0.429 | 0.2506 | Yes |
| 18 | Sirt1 | 3017 | 0.428 | 0.2633 | Yes |
| 19 | Nefh | 3364 | 0.403 | 0.2634 | Yes |
| 20 | Pgs1 | 3428 | 0.399 | 0.2736 | Yes |
| 21 | Pomc | 3479 | 0.398 | 0.2842 | Yes |
| 22 | Alox15 | 3710 | 0.386 | 0.2879 | Yes |
| 23 | She | 3826 | 0.379 | 0.2956 | Yes |
| 24 | Ddx4 | 4174 | 0.361 | 0.2943 | Yes |
| 25 | Papolb | 4484 | 0.346 | 0.2939 | Yes |
| 26 | Nphp1 | 4683 | 0.339 | 0.2973 | Yes |
| 27 | Ncaph | 4751 | 0.336 | 0.3054 | Yes |
| 28 | Oaz3 | 4990 | 0.325 | 0.3069 | Yes |
| 29 | Jam3 | 5874 | 0.288 | 0.2838 | Yes |
| 30 | Aurka | 5957 | 0.284 | 0.2897 | Yes |
| 31 | Slc2a5 | 6049 | 0.280 | 0.2952 | Yes |
| 32 | Mlf1 | 6052 | 0.280 | 0.3039 | Yes |
| 33 | Prkar2a | 6365 | 0.270 | 0.3010 | Yes |
| 34 | Ttk | 6389 | 0.269 | 0.3086 | Yes |
| 35 | Ide | 6396 | 0.269 | 0.3168 | Yes |
| 36 | Rfc4 | 6536 | 0.261 | 0.3199 | Yes |
| 37 | Dcc | 6569 | 0.259 | 0.3268 | Yes |
| 38 | Naa11 | 7031 | 0.238 | 0.3175 | No |
| 39 | Agfg1 | 7624 | 0.220 | 0.3029 | No |
| 40 | Dbf4 | 7731 | 0.217 | 0.3058 | No |
| 41 | Ace | 8183 | 0.201 | 0.2957 | No |
| 42 | Clgn | 8324 | 0.196 | 0.2967 | No |
| 43 | Lpin1 | 8675 | 0.182 | 0.2897 | No |
| 44 | Gpr182 | 8835 | 0.175 | 0.2894 | No |
| 45 | Vdac3 | 8859 | 0.174 | 0.2940 | No |
| 46 | Pias2 | 8887 | 0.173 | 0.2985 | No |
| 47 | Scg3 | 8971 | 0.170 | 0.3008 | No |
| 48 | Nf2 | 9051 | 0.166 | 0.3031 | No |
| 49 | Zc3h14 | 9372 | 0.160 | 0.2965 | No |
| 50 | Adam2 | 9847 | 0.141 | 0.2837 | No |
| 51 | Parp2 | 10132 | 0.138 | 0.2777 | No |
| 52 | Strbp | 10145 | 0.138 | 0.2816 | No |
| 53 | Arl4a | 10243 | 0.134 | 0.2822 | No |
| 54 | Chfr | 10388 | 0.129 | 0.2810 | No |
| 55 | Ccnb2 | 10527 | 0.124 | 0.2799 | No |
| 56 | Tulp2 | 10562 | 0.122 | 0.2824 | No |
| 57 | Clpb | 10645 | 0.118 | 0.2832 | No |
| 58 | Chrm4 | 10670 | 0.118 | 0.2860 | No |
| 59 | Ift88 | 10725 | 0.116 | 0.2877 | No |
| 60 | Ip6k1 | 11117 | 0.102 | 0.2766 | No |
| 61 | Mep1b | 11232 | 0.097 | 0.2755 | No |
| 62 | Mast2 | 11295 | 0.095 | 0.2762 | No |
| 63 | Rad17 | 11296 | 0.095 | 0.2792 | No |
| 64 | Ezh2 | 11297 | 0.095 | 0.2822 | No |
| 65 | Il12rb2 | 12343 | 0.056 | 0.2459 | No |
| 66 | Csnk2a2 | 12571 | 0.048 | 0.2392 | No |
| 67 | Scg5 | 12620 | 0.046 | 0.2389 | No |
| 68 | Mtor | 12809 | 0.039 | 0.2333 | No |
| 69 | Slc12a2 | 12854 | 0.037 | 0.2328 | No |
| 70 | Actl7b | 13396 | 0.019 | 0.2137 | No |
| 71 | Dnajb8 | 13570 | 0.014 | 0.2079 | No |
| 72 | Gmcl1 | 13622 | 0.012 | 0.2064 | No |
| 73 | Tle4 | 14036 | 0.001 | 0.1914 | No |
| 74 | Il13ra2 | 14089 | 0.000 | 0.1895 | No |
| 75 | Tssk2 | 14109 | 0.000 | 0.1888 | No |
| 76 | Crisp2 | 14774 | -0.014 | 0.1651 | No |
| 77 | Stam2 | 14777 | -0.014 | 0.1655 | No |
| 78 | Dmrt1 | 14866 | -0.017 | 0.1628 | No |
| 79 | Tsn | 15014 | -0.023 | 0.1582 | No |
| 80 | Cftr | 15308 | -0.033 | 0.1486 | No |
| 81 | Pacrg | 15498 | -0.040 | 0.1429 | No |
| 82 | Camk4 | 15669 | -0.046 | 0.1382 | No |
| 83 | Acrv1 | 16194 | -0.064 | 0.1212 | No |
| 84 | Tnni3 | 16367 | -0.070 | 0.1171 | No |
| 85 | Coil | 16483 | -0.075 | 0.1153 | No |
| 86 | Dmc1 | 17124 | -0.097 | 0.0950 | No |
| 87 | Mllt10 | 17374 | -0.106 | 0.0893 | No |
| 88 | Grm8 | 17663 | -0.116 | 0.0824 | No |
| 89 | Braf | 17764 | -0.120 | 0.0825 | No |
| 90 | Ldhc | 17950 | -0.126 | 0.0798 | No |
| 91 | Pebp1 | 17982 | -0.127 | 0.0826 | No |
| 92 | Clvs1 | 18066 | -0.130 | 0.0837 | No |
| 93 | Phkg2 | 18415 | -0.144 | 0.0755 | No |
| 94 | Nos1 | 18655 | -0.152 | 0.0716 | No |
| 95 | Cnih2 | 19106 | -0.167 | 0.0605 | No |
| 96 | Hspa2 | 19331 | -0.176 | 0.0578 | No |
| 97 | Cdkn3 | 19384 | -0.178 | 0.0615 | No |
| 98 | Ddx25 | 20008 | -0.200 | 0.0451 | No |
| 99 | Art3 | 20108 | -0.204 | 0.0479 | No |
| 100 | Map7 | 20508 | -0.219 | 0.0402 | No |
| 101 | Adad1 | 21021 | -0.233 | 0.0289 | No |
| 102 | Gstm5 | 22442 | -0.292 | -0.0136 | No |
| 103 | Psmg1 | 22461 | -0.293 | -0.0051 | No |
| 104 | Ybx2 | 23034 | -0.318 | -0.0159 | No |
| 105 | Phf7 | 23036 | -0.318 | -0.0060 | No |
| 106 | Tcp11 | 23744 | -0.357 | -0.0205 | No |
| 107 | Gad1 | 25144 | -0.444 | -0.0575 | No |
| 108 | Taldo1 | 25656 | -0.489 | -0.0608 | No |
| 109 | Spata6 | 25943 | -0.520 | -0.0549 | No |
| 110 | Cct6b | 26201 | -0.547 | -0.0471 | No |
| 111 | Zpbp | 26474 | -0.588 | -0.0386 | No |
| 112 | Rpl39l | 26592 | -0.604 | -0.0239 | No |
| 113 | Gapdhs | 26624 | -0.609 | -0.0059 | No |
| 114 | Acrbp | 26881 | -0.655 | 0.0053 | No |
| 115 | Pcsk4 | 26951 | -0.668 | 0.0237 | No |