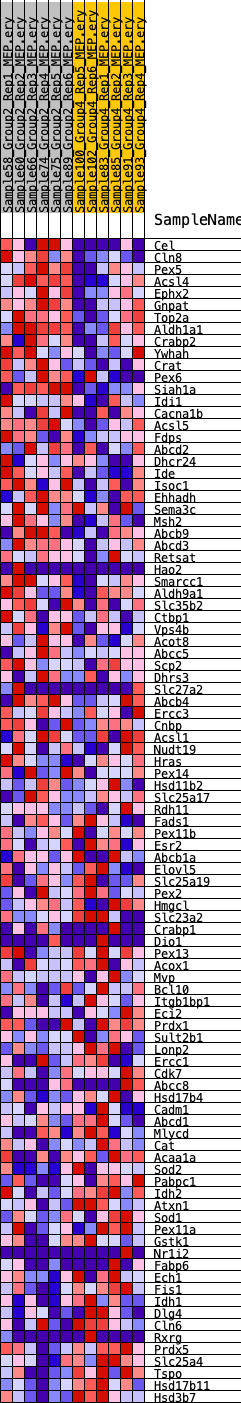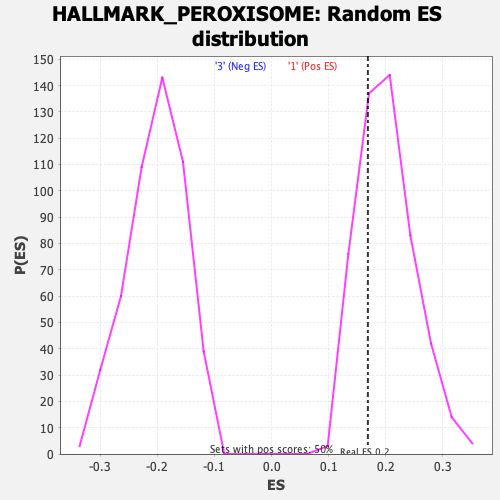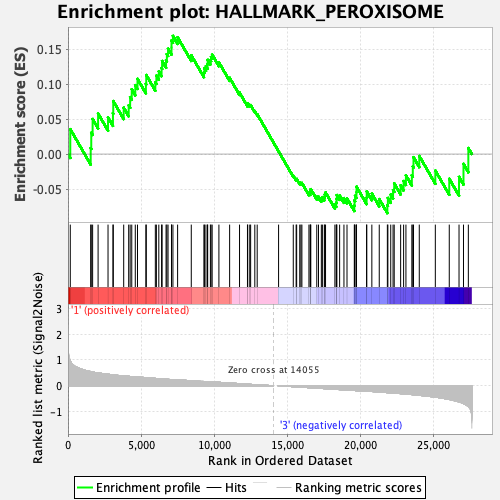
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.16893572 |
| Normalized Enrichment Score (NES) | 0.83796823 |
| Nominal p-value | 0.7196819 |
| FDR q-value | 0.7164152 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cel | 156 | 0.958 | 0.0356 | Yes |
| 2 | Cln8 | 1551 | 0.549 | 0.0085 | Yes |
| 3 | Pex5 | 1590 | 0.544 | 0.0306 | Yes |
| 4 | Acsl4 | 1683 | 0.532 | 0.0501 | Yes |
| 5 | Ephx2 | 2057 | 0.497 | 0.0580 | Yes |
| 6 | Gnpat | 2736 | 0.444 | 0.0524 | Yes |
| 7 | Top2a | 3073 | 0.422 | 0.0584 | Yes |
| 8 | Aldh1a1 | 3094 | 0.421 | 0.0758 | Yes |
| 9 | Crabp2 | 3807 | 0.381 | 0.0663 | Yes |
| 10 | Ywhah | 4148 | 0.364 | 0.0696 | Yes |
| 11 | Crat | 4247 | 0.358 | 0.0814 | Yes |
| 12 | Pex6 | 4358 | 0.352 | 0.0925 | Yes |
| 13 | Siah1a | 4599 | 0.341 | 0.0985 | Yes |
| 14 | Idi1 | 4750 | 0.336 | 0.1075 | Yes |
| 15 | Cacna1b | 5323 | 0.318 | 0.1003 | Yes |
| 16 | Acsl5 | 5350 | 0.316 | 0.1130 | Yes |
| 17 | Fdps | 5969 | 0.283 | 0.1027 | Yes |
| 18 | Abcd2 | 6042 | 0.280 | 0.1122 | Yes |
| 19 | Dhcr24 | 6207 | 0.276 | 0.1181 | Yes |
| 20 | Ide | 6396 | 0.269 | 0.1228 | Yes |
| 21 | Isoc1 | 6432 | 0.267 | 0.1330 | Yes |
| 22 | Ehhadh | 6703 | 0.253 | 0.1341 | Yes |
| 23 | Sema3c | 6758 | 0.250 | 0.1429 | Yes |
| 24 | Msh2 | 6838 | 0.247 | 0.1506 | Yes |
| 25 | Abcb9 | 7074 | 0.236 | 0.1522 | Yes |
| 26 | Abcd3 | 7075 | 0.236 | 0.1624 | Yes |
| 27 | Retsat | 7172 | 0.233 | 0.1689 | Yes |
| 28 | Hao2 | 7492 | 0.223 | 0.1669 | No |
| 29 | Smarcc1 | 8426 | 0.191 | 0.1412 | No |
| 30 | Aldh9a1 | 9292 | 0.163 | 0.1168 | No |
| 31 | Slc35b2 | 9324 | 0.162 | 0.1227 | No |
| 32 | Ctbp1 | 9430 | 0.158 | 0.1256 | No |
| 33 | Vps4b | 9536 | 0.154 | 0.1284 | No |
| 34 | Acot8 | 9547 | 0.153 | 0.1346 | No |
| 35 | Abcc5 | 9733 | 0.147 | 0.1342 | No |
| 36 | Scp2 | 9778 | 0.145 | 0.1388 | No |
| 37 | Dhrs3 | 9853 | 0.141 | 0.1422 | No |
| 38 | Slc27a2 | 10315 | 0.131 | 0.1311 | No |
| 39 | Abcb4 | 11048 | 0.104 | 0.1090 | No |
| 40 | Ercc3 | 11725 | 0.080 | 0.0878 | No |
| 41 | Cnbp | 12267 | 0.059 | 0.0707 | No |
| 42 | Acsl1 | 12285 | 0.059 | 0.0726 | No |
| 43 | Nudt19 | 12424 | 0.053 | 0.0699 | No |
| 44 | Hras | 12484 | 0.051 | 0.0700 | No |
| 45 | Pex14 | 12769 | 0.040 | 0.0614 | No |
| 46 | Hsd11b2 | 12932 | 0.034 | 0.0570 | No |
| 47 | Slc25a17 | 14394 | -0.001 | 0.0039 | No |
| 48 | Rdh11 | 15398 | -0.036 | -0.0310 | No |
| 49 | Fads1 | 15594 | -0.043 | -0.0362 | No |
| 50 | Pex11b | 15630 | -0.045 | -0.0356 | No |
| 51 | Esr2 | 15841 | -0.052 | -0.0409 | No |
| 52 | Abcb1a | 15919 | -0.054 | -0.0414 | No |
| 53 | Elovl5 | 15987 | -0.058 | -0.0414 | No |
| 54 | Slc25a19 | 16469 | -0.074 | -0.0557 | No |
| 55 | Pex2 | 16553 | -0.077 | -0.0554 | No |
| 56 | Hmgcl | 16574 | -0.077 | -0.0528 | No |
| 57 | Slc23a2 | 16596 | -0.078 | -0.0502 | No |
| 58 | Crabp1 | 17004 | -0.092 | -0.0610 | No |
| 59 | Dio1 | 17122 | -0.097 | -0.0611 | No |
| 60 | Pex13 | 17316 | -0.104 | -0.0636 | No |
| 61 | Acox1 | 17388 | -0.107 | -0.0616 | No |
| 62 | Mvp | 17520 | -0.110 | -0.0616 | No |
| 63 | Bcl10 | 17549 | -0.112 | -0.0578 | No |
| 64 | Itgb1bp1 | 17604 | -0.114 | -0.0549 | No |
| 65 | Eci2 | 18243 | -0.137 | -0.0722 | No |
| 66 | Prdx1 | 18334 | -0.141 | -0.0694 | No |
| 67 | Sult2b1 | 18358 | -0.142 | -0.0641 | No |
| 68 | Lonp2 | 18375 | -0.142 | -0.0586 | No |
| 69 | Ercc1 | 18567 | -0.149 | -0.0591 | No |
| 70 | Cdk7 | 18862 | -0.159 | -0.0630 | No |
| 71 | Abcc8 | 19071 | -0.166 | -0.0634 | No |
| 72 | Hsd17b4 | 19570 | -0.184 | -0.0736 | No |
| 73 | Cadm1 | 19585 | -0.185 | -0.0661 | No |
| 74 | Abcd1 | 19624 | -0.186 | -0.0595 | No |
| 75 | Mlycd | 19717 | -0.190 | -0.0547 | No |
| 76 | Cat | 19721 | -0.190 | -0.0466 | No |
| 77 | Acaa1a | 20411 | -0.215 | -0.0624 | No |
| 78 | Sod2 | 20419 | -0.215 | -0.0534 | No |
| 79 | Pabpc1 | 20769 | -0.223 | -0.0565 | No |
| 80 | Idh2 | 21275 | -0.244 | -0.0644 | No |
| 81 | Atxn1 | 21829 | -0.267 | -0.0730 | No |
| 82 | Sod1 | 21865 | -0.268 | -0.0627 | No |
| 83 | Pex11a | 22060 | -0.276 | -0.0579 | No |
| 84 | Gstk1 | 22219 | -0.284 | -0.0514 | No |
| 85 | Nr1i2 | 22298 | -0.287 | -0.0419 | No |
| 86 | Fabp6 | 22737 | -0.305 | -0.0447 | No |
| 87 | Ech1 | 22940 | -0.314 | -0.0386 | No |
| 88 | Fis1 | 23099 | -0.322 | -0.0305 | No |
| 89 | Idh1 | 23503 | -0.342 | -0.0304 | No |
| 90 | Dlg4 | 23569 | -0.347 | -0.0178 | No |
| 91 | Cln6 | 23617 | -0.349 | -0.0045 | No |
| 92 | Rxrg | 24011 | -0.372 | -0.0028 | No |
| 93 | Prdx5 | 25108 | -0.441 | -0.0236 | No |
| 94 | Slc25a4 | 26061 | -0.533 | -0.0353 | No |
| 95 | Tspo | 26728 | -0.625 | -0.0326 | No |
| 96 | Hsd17b11 | 27033 | -0.688 | -0.0141 | No |
| 97 | Hsd3b7 | 27365 | -0.806 | 0.0086 | No |