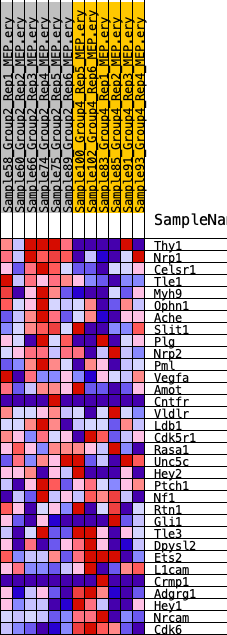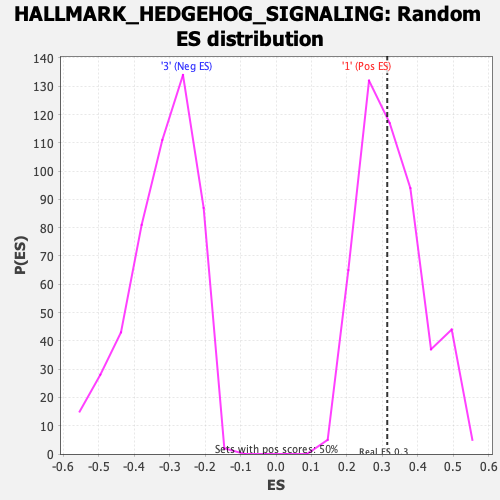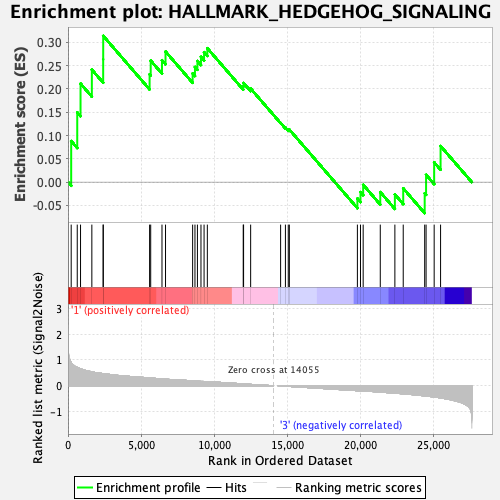
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.31390074 |
| Normalized Enrichment Score (NES) | 0.96527433 |
| Nominal p-value | 0.49098197 |
| FDR q-value | 0.7472452 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thy1 | 218 | 0.892 | 0.0882 | Yes |
| 2 | Nrp1 | 632 | 0.710 | 0.1496 | Yes |
| 3 | Celsr1 | 857 | 0.650 | 0.2114 | Yes |
| 4 | Tle1 | 1630 | 0.538 | 0.2414 | Yes |
| 5 | Myh9 | 2405 | 0.467 | 0.2636 | Yes |
| 6 | Ophn1 | 2407 | 0.467 | 0.3139 | Yes |
| 7 | Ache | 5578 | 0.301 | 0.2314 | No |
| 8 | Slit1 | 5653 | 0.298 | 0.2608 | No |
| 9 | Plg | 6424 | 0.267 | 0.2616 | No |
| 10 | Nrp2 | 6666 | 0.255 | 0.2803 | No |
| 11 | Pml | 8519 | 0.188 | 0.2334 | No |
| 12 | Vegfa | 8678 | 0.182 | 0.2472 | No |
| 13 | Amot | 8846 | 0.175 | 0.2600 | No |
| 14 | Cntfr | 9089 | 0.165 | 0.2690 | No |
| 15 | Vldlr | 9301 | 0.163 | 0.2789 | No |
| 16 | Ldb1 | 9524 | 0.154 | 0.2874 | No |
| 17 | Cdk5r1 | 11977 | 0.071 | 0.2061 | No |
| 18 | Rasa1 | 12002 | 0.070 | 0.2127 | No |
| 19 | Unc5c | 12485 | 0.051 | 0.2008 | No |
| 20 | Hey2 | 14533 | -0.005 | 0.1270 | No |
| 21 | Ptch1 | 14861 | -0.017 | 0.1170 | No |
| 22 | Nf1 | 15056 | -0.024 | 0.1126 | No |
| 23 | Rtn1 | 15130 | -0.026 | 0.1128 | No |
| 24 | Gli1 | 19781 | -0.192 | -0.0352 | No |
| 25 | Tle3 | 19993 | -0.199 | -0.0214 | No |
| 26 | Dpysl2 | 20176 | -0.207 | -0.0058 | No |
| 27 | Ets2 | 21345 | -0.248 | -0.0214 | No |
| 28 | L1cam | 22343 | -0.289 | -0.0265 | No |
| 29 | Crmp1 | 22914 | -0.314 | -0.0134 | No |
| 30 | Adgrg1 | 24381 | -0.395 | -0.0241 | No |
| 31 | Hey1 | 24476 | -0.402 | 0.0157 | No |
| 32 | Nrcam | 25029 | -0.435 | 0.0425 | No |
| 33 | Cdk6 | 25467 | -0.472 | 0.0774 | No |