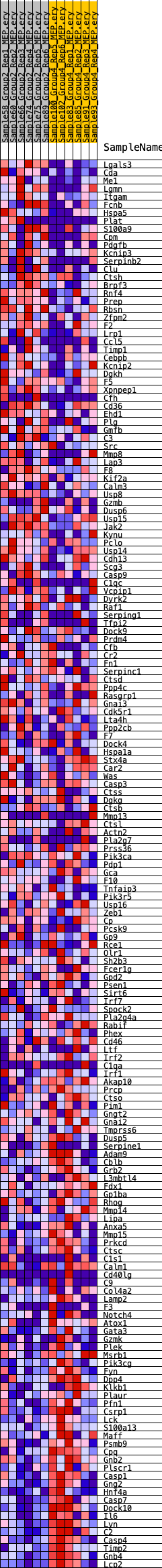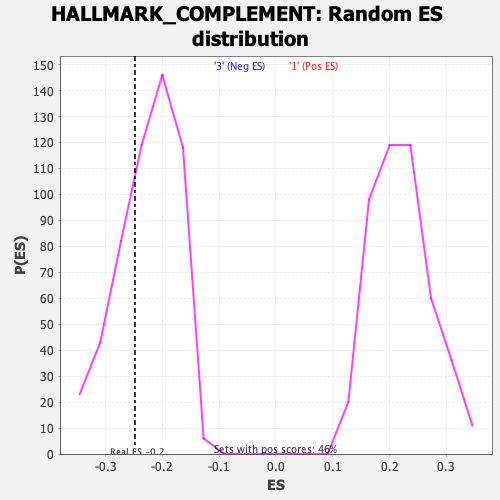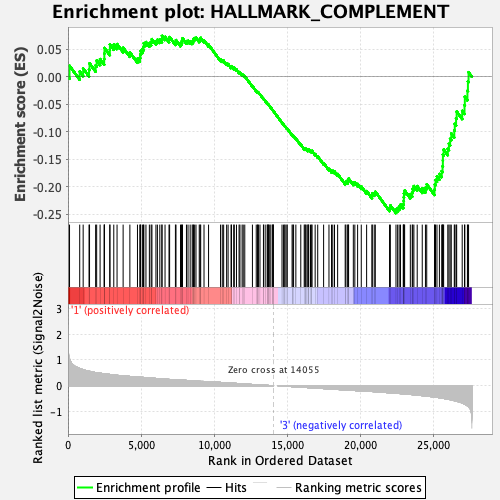
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.24829982 |
| Normalized Enrichment Score (NES) | -1.102233 |
| Nominal p-value | 0.31843576 |
| FDR q-value | 0.61589485 |
| FWER p-Value | 0.97 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lgals3 | 104 | 1.046 | 0.0198 | No |
| 2 | Cda | 800 | 0.665 | 0.0094 | No |
| 3 | Me1 | 1036 | 0.621 | 0.0149 | No |
| 4 | Lgmn | 1438 | 0.561 | 0.0129 | No |
| 5 | Itgam | 1472 | 0.558 | 0.0243 | No |
| 6 | Fcnb | 1889 | 0.510 | 0.0206 | No |
| 7 | Hspa5 | 1959 | 0.504 | 0.0295 | No |
| 8 | Plat | 2193 | 0.488 | 0.0320 | No |
| 9 | S100a9 | 2473 | 0.461 | 0.0322 | No |
| 10 | Cpm | 2484 | 0.460 | 0.0422 | No |
| 11 | Pdgfb | 2493 | 0.459 | 0.0523 | No |
| 12 | Kcnip3 | 2859 | 0.438 | 0.0488 | No |
| 13 | Serpinb2 | 2864 | 0.437 | 0.0585 | No |
| 14 | Clu | 3129 | 0.418 | 0.0583 | No |
| 15 | Ctsh | 3353 | 0.404 | 0.0593 | No |
| 16 | Brpf3 | 3768 | 0.383 | 0.0529 | No |
| 17 | Rnf4 | 4229 | 0.359 | 0.0442 | No |
| 18 | Prep | 4748 | 0.336 | 0.0329 | No |
| 19 | Rbsn | 4912 | 0.328 | 0.0343 | No |
| 20 | Zfpm2 | 4942 | 0.326 | 0.0406 | No |
| 21 | F2 | 4967 | 0.326 | 0.0471 | No |
| 22 | Lrp1 | 5085 | 0.323 | 0.0501 | No |
| 23 | Ccl5 | 5160 | 0.321 | 0.0547 | No |
| 24 | Timp1 | 5185 | 0.320 | 0.0610 | No |
| 25 | Cebpb | 5328 | 0.317 | 0.0630 | No |
| 26 | Kcnip2 | 5562 | 0.302 | 0.0613 | No |
| 27 | Dgkh | 5682 | 0.297 | 0.0636 | No |
| 28 | F5 | 5737 | 0.294 | 0.0683 | No |
| 29 | Xpnpep1 | 6011 | 0.281 | 0.0647 | No |
| 30 | Cfh | 6111 | 0.277 | 0.0673 | No |
| 31 | Cd36 | 6276 | 0.273 | 0.0675 | No |
| 32 | Ehd1 | 6419 | 0.267 | 0.0684 | No |
| 33 | Plg | 6424 | 0.267 | 0.0743 | No |
| 34 | Gmfb | 6633 | 0.256 | 0.0725 | No |
| 35 | C3 | 6905 | 0.244 | 0.0681 | No |
| 36 | Src | 6946 | 0.242 | 0.0721 | No |
| 37 | Mmp8 | 7342 | 0.229 | 0.0628 | No |
| 38 | Lap3 | 7381 | 0.227 | 0.0666 | No |
| 39 | F8 | 7690 | 0.219 | 0.0603 | No |
| 40 | Kif2a | 7757 | 0.215 | 0.0627 | No |
| 41 | Calm3 | 7795 | 0.214 | 0.0662 | No |
| 42 | Usp8 | 7826 | 0.213 | 0.0699 | No |
| 43 | Gzmb | 8097 | 0.205 | 0.0647 | No |
| 44 | Dusp6 | 8203 | 0.200 | 0.0653 | No |
| 45 | Usp15 | 8350 | 0.194 | 0.0644 | No |
| 46 | Jak2 | 8488 | 0.189 | 0.0637 | No |
| 47 | Kynu | 8548 | 0.187 | 0.0657 | No |
| 48 | Pclo | 8580 | 0.186 | 0.0688 | No |
| 49 | Usp14 | 8656 | 0.183 | 0.0702 | No |
| 50 | Cdh13 | 8748 | 0.178 | 0.0709 | No |
| 51 | Scg3 | 8971 | 0.170 | 0.0666 | No |
| 52 | Casp9 | 9029 | 0.167 | 0.0683 | No |
| 53 | C1qc | 9070 | 0.166 | 0.0706 | No |
| 54 | Vcpip1 | 9297 | 0.163 | 0.0661 | No |
| 55 | Dyrk2 | 9608 | 0.151 | 0.0582 | No |
| 56 | Raf1 | 10427 | 0.127 | 0.0312 | No |
| 57 | Serping1 | 10545 | 0.123 | 0.0297 | No |
| 58 | Tfpi2 | 10639 | 0.119 | 0.0290 | No |
| 59 | Dock9 | 10843 | 0.112 | 0.0241 | No |
| 60 | Prdm4 | 10943 | 0.108 | 0.0229 | No |
| 61 | Cfb | 11145 | 0.101 | 0.0179 | No |
| 62 | Cr2 | 11175 | 0.099 | 0.0190 | No |
| 63 | Fn1 | 11328 | 0.094 | 0.0156 | No |
| 64 | Serpinc1 | 11366 | 0.093 | 0.0164 | No |
| 65 | Ctsd | 11515 | 0.088 | 0.0129 | No |
| 66 | Ppp4c | 11693 | 0.081 | 0.0083 | No |
| 67 | Rasgrp1 | 11753 | 0.079 | 0.0080 | No |
| 68 | Gnai3 | 11901 | 0.073 | 0.0043 | No |
| 69 | Cdk5r1 | 11977 | 0.071 | 0.0031 | No |
| 70 | Lta4h | 12083 | 0.066 | 0.0008 | No |
| 71 | Ppp2cb | 12606 | 0.047 | -0.0172 | No |
| 72 | F7 | 12867 | 0.037 | -0.0258 | No |
| 73 | Dock4 | 12961 | 0.033 | -0.0285 | No |
| 74 | Hspa1a | 12969 | 0.033 | -0.0280 | No |
| 75 | Stx4a | 13015 | 0.032 | -0.0289 | No |
| 76 | Car2 | 13122 | 0.028 | -0.0322 | No |
| 77 | Was | 13359 | 0.020 | -0.0403 | No |
| 78 | Casp3 | 13385 | 0.020 | -0.0408 | No |
| 79 | Ctss | 13523 | 0.016 | -0.0454 | No |
| 80 | Dgkg | 13653 | 0.011 | -0.0499 | No |
| 81 | Ctsb | 13657 | 0.011 | -0.0497 | No |
| 82 | Mmp13 | 13676 | 0.011 | -0.0501 | No |
| 83 | Ctsl | 13713 | 0.010 | -0.0512 | No |
| 84 | Actn2 | 13790 | 0.007 | -0.0538 | No |
| 85 | Pla2g7 | 13867 | 0.004 | -0.0565 | No |
| 86 | Prss36 | 13993 | 0.002 | -0.0610 | No |
| 87 | Pik3ca | 14002 | 0.002 | -0.0613 | No |
| 88 | Pdp1 | 14026 | 0.001 | -0.0621 | No |
| 89 | Gca | 14032 | 0.001 | -0.0622 | No |
| 90 | F10 | 14629 | -0.009 | -0.0838 | No |
| 91 | Tnfaip3 | 14735 | -0.013 | -0.0873 | No |
| 92 | Pik3r5 | 14823 | -0.016 | -0.0901 | No |
| 93 | Usp16 | 14832 | -0.016 | -0.0901 | No |
| 94 | Zeb1 | 14969 | -0.021 | -0.0946 | No |
| 95 | Cp | 14976 | -0.021 | -0.0943 | No |
| 96 | Pcsk9 | 15317 | -0.033 | -0.1059 | No |
| 97 | Gp9 | 15375 | -0.036 | -0.1072 | No |
| 98 | Rce1 | 15419 | -0.037 | -0.1079 | No |
| 99 | Olr1 | 15575 | -0.043 | -0.1126 | No |
| 100 | Sh2b3 | 15911 | -0.054 | -0.1236 | No |
| 101 | Fcer1g | 16147 | -0.063 | -0.1308 | No |
| 102 | Gpd2 | 16212 | -0.065 | -0.1316 | No |
| 103 | Psen1 | 16215 | -0.065 | -0.1303 | No |
| 104 | Sirt6 | 16329 | -0.069 | -0.1328 | No |
| 105 | Irf7 | 16405 | -0.072 | -0.1339 | No |
| 106 | Spock2 | 16418 | -0.072 | -0.1327 | No |
| 107 | Pla2g4a | 16454 | -0.073 | -0.1324 | No |
| 108 | Rabif | 16576 | -0.077 | -0.1350 | No |
| 109 | Phex | 16662 | -0.081 | -0.1363 | No |
| 110 | Cd46 | 16673 | -0.081 | -0.1348 | No |
| 111 | Ltf | 16894 | -0.089 | -0.1409 | No |
| 112 | Irf2 | 17069 | -0.095 | -0.1451 | No |
| 113 | C1qa | 17472 | -0.110 | -0.1573 | No |
| 114 | Irf1 | 17838 | -0.122 | -0.1678 | No |
| 115 | Akap10 | 18013 | -0.128 | -0.1713 | No |
| 116 | Prcp | 18098 | -0.131 | -0.1714 | No |
| 117 | Ctso | 18217 | -0.136 | -0.1726 | No |
| 118 | Pim1 | 18431 | -0.144 | -0.1771 | No |
| 119 | Gngt2 | 18947 | -0.162 | -0.1922 | No |
| 120 | Gnai2 | 18997 | -0.163 | -0.1904 | No |
| 121 | Tmprss6 | 19120 | -0.168 | -0.1910 | No |
| 122 | Dusp5 | 19142 | -0.169 | -0.1880 | No |
| 123 | Serpine1 | 19179 | -0.170 | -0.1855 | No |
| 124 | Adam9 | 19503 | -0.182 | -0.1931 | No |
| 125 | Cblb | 19606 | -0.185 | -0.1927 | No |
| 126 | Grb2 | 19796 | -0.192 | -0.1952 | No |
| 127 | L3mbtl4 | 20046 | -0.202 | -0.1998 | No |
| 128 | Fdx1 | 20412 | -0.215 | -0.2082 | No |
| 129 | Gp1ba | 20776 | -0.223 | -0.2164 | No |
| 130 | Rhog | 20797 | -0.224 | -0.2121 | No |
| 131 | Mmp14 | 20931 | -0.229 | -0.2118 | No |
| 132 | Lipa | 21005 | -0.233 | -0.2092 | No |
| 133 | Anxa5 | 21978 | -0.273 | -0.2385 | No |
| 134 | Mmp15 | 22034 | -0.275 | -0.2343 | No |
| 135 | Prkcd | 22419 | -0.291 | -0.2417 | Yes |
| 136 | Ctsc | 22532 | -0.296 | -0.2391 | Yes |
| 137 | C1s1 | 22636 | -0.300 | -0.2361 | Yes |
| 138 | Calm1 | 22729 | -0.305 | -0.2326 | Yes |
| 139 | Cd40lg | 22915 | -0.314 | -0.2323 | Yes |
| 140 | C9 | 22947 | -0.314 | -0.2263 | Yes |
| 141 | Col4a2 | 22954 | -0.314 | -0.2195 | Yes |
| 142 | Lamp2 | 22964 | -0.315 | -0.2127 | Yes |
| 143 | F3 | 23010 | -0.317 | -0.2072 | Yes |
| 144 | Notch4 | 23399 | -0.337 | -0.2137 | Yes |
| 145 | Atox1 | 23529 | -0.344 | -0.2107 | Yes |
| 146 | Gata3 | 23562 | -0.346 | -0.2040 | Yes |
| 147 | Gzmk | 23642 | -0.350 | -0.1990 | Yes |
| 148 | Plek | 23870 | -0.364 | -0.1991 | Yes |
| 149 | Msrb1 | 24214 | -0.384 | -0.2029 | Yes |
| 150 | Pik3cg | 24433 | -0.398 | -0.2019 | Yes |
| 151 | Fyn | 24520 | -0.404 | -0.1959 | Yes |
| 152 | Dpp4 | 25051 | -0.436 | -0.2054 | Yes |
| 153 | Klkb1 | 25071 | -0.438 | -0.1962 | Yes |
| 154 | Plaur | 25120 | -0.442 | -0.1880 | Yes |
| 155 | Pfn1 | 25220 | -0.450 | -0.1815 | Yes |
| 156 | Csrp1 | 25387 | -0.464 | -0.1770 | Yes |
| 157 | Lck | 25545 | -0.480 | -0.1719 | Yes |
| 158 | S100a13 | 25603 | -0.485 | -0.1631 | Yes |
| 159 | Maff | 25623 | -0.486 | -0.1528 | Yes |
| 160 | Psmb9 | 25627 | -0.487 | -0.1420 | Yes |
| 161 | Cpq | 25676 | -0.491 | -0.1326 | Yes |
| 162 | Gnb2 | 25961 | -0.522 | -0.1312 | Yes |
| 163 | Plscr1 | 26038 | -0.531 | -0.1220 | Yes |
| 164 | Casp1 | 26120 | -0.539 | -0.1128 | Yes |
| 165 | Gng2 | 26197 | -0.547 | -0.1033 | Yes |
| 166 | Hnf4a | 26404 | -0.578 | -0.0977 | Yes |
| 167 | Casp7 | 26437 | -0.583 | -0.0858 | Yes |
| 168 | Dock10 | 26527 | -0.595 | -0.0756 | Yes |
| 169 | Il6 | 26567 | -0.602 | -0.0635 | Yes |
| 170 | Lyn | 26945 | -0.667 | -0.0622 | Yes |
| 171 | C2 | 27103 | -0.708 | -0.0519 | Yes |
| 172 | Casp4 | 27125 | -0.716 | -0.0366 | Yes |
| 173 | Timp2 | 27302 | -0.778 | -0.0254 | Yes |
| 174 | Gnb4 | 27336 | -0.792 | -0.0088 | Yes |
| 175 | Lcp2 | 27380 | -0.818 | 0.0081 | Yes |