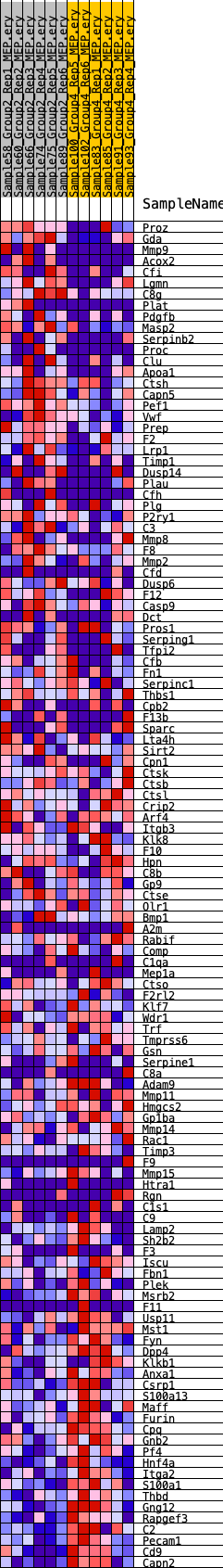
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
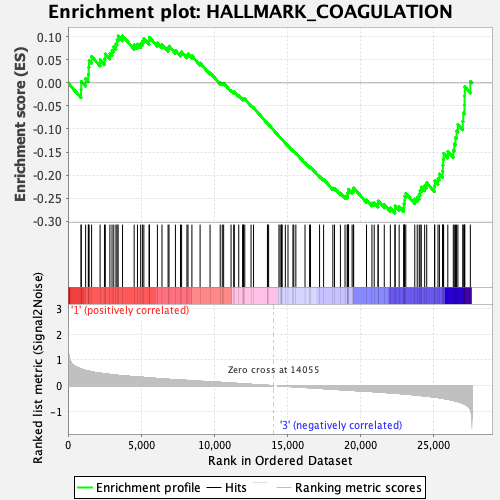
| GeneSet | HALLMARK_COAGULATION |
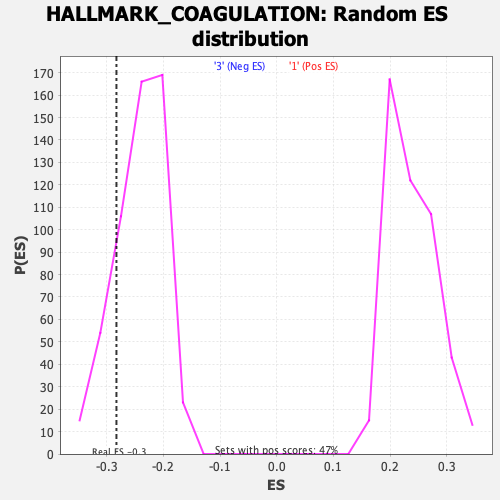
| Enrichment Score (ES) | -0.28269833 |
| Normalized Enrichment Score (NES) | -1.169752 |
| Nominal p-value | 0.1575985 |
| FDR q-value | 0.7531234 |
| FWER p-Value | 0.944 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Proz | 892 | 0.643 | -0.0143 | No |
| 2 | Gda | 903 | 0.641 | 0.0035 | No |
| 3 | Mmp9 | 1205 | 0.592 | 0.0093 | No |
| 4 | Acox2 | 1388 | 0.567 | 0.0187 | No |
| 5 | Cfi | 1412 | 0.564 | 0.0338 | No |
| 6 | Lgmn | 1438 | 0.561 | 0.0487 | No |
| 7 | C8g | 1613 | 0.540 | 0.0577 | No |
| 8 | Plat | 2193 | 0.488 | 0.0504 | No |
| 9 | Pdgfb | 2493 | 0.459 | 0.0525 | No |
| 10 | Masp2 | 2550 | 0.454 | 0.0633 | No |
| 11 | Serpinb2 | 2864 | 0.437 | 0.0642 | No |
| 12 | Proc | 3009 | 0.428 | 0.0711 | No |
| 13 | Clu | 3129 | 0.418 | 0.0786 | No |
| 14 | Apoa1 | 3278 | 0.409 | 0.0848 | No |
| 15 | Ctsh | 3353 | 0.404 | 0.0935 | No |
| 16 | Capn5 | 3426 | 0.399 | 0.1022 | No |
| 17 | Pef1 | 3728 | 0.386 | 0.1021 | No |
| 18 | Vwf | 4522 | 0.345 | 0.0830 | No |
| 19 | Prep | 4748 | 0.336 | 0.0843 | No |
| 20 | F2 | 4967 | 0.326 | 0.0856 | No |
| 21 | Lrp1 | 5085 | 0.323 | 0.0905 | No |
| 22 | Timp1 | 5185 | 0.320 | 0.0959 | No |
| 23 | Dusp14 | 5539 | 0.304 | 0.0916 | No |
| 24 | Plau | 5568 | 0.302 | 0.0991 | No |
| 25 | Cfh | 6111 | 0.277 | 0.0873 | No |
| 26 | Plg | 6424 | 0.267 | 0.0835 | No |
| 27 | P2ry1 | 6841 | 0.247 | 0.0753 | No |
| 28 | C3 | 6905 | 0.244 | 0.0799 | No |
| 29 | Mmp8 | 7342 | 0.229 | 0.0705 | No |
| 30 | F8 | 7690 | 0.219 | 0.0640 | No |
| 31 | Mmp2 | 7764 | 0.215 | 0.0675 | No |
| 32 | Cfd | 8129 | 0.203 | 0.0600 | No |
| 33 | Dusp6 | 8203 | 0.200 | 0.0630 | No |
| 34 | F12 | 8469 | 0.190 | 0.0587 | No |
| 35 | Casp9 | 9029 | 0.167 | 0.0431 | No |
| 36 | Dct | 9710 | 0.147 | 0.0225 | No |
| 37 | Pros1 | 10405 | 0.128 | 0.0009 | No |
| 38 | Serping1 | 10545 | 0.123 | -0.0007 | No |
| 39 | Tfpi2 | 10639 | 0.119 | -0.0007 | No |
| 40 | Cfb | 11145 | 0.101 | -0.0163 | No |
| 41 | Fn1 | 11328 | 0.094 | -0.0202 | No |
| 42 | Serpinc1 | 11366 | 0.093 | -0.0190 | No |
| 43 | Thbs1 | 11671 | 0.082 | -0.0277 | No |
| 44 | Cpb2 | 11933 | 0.072 | -0.0351 | No |
| 45 | F13b | 11986 | 0.070 | -0.0350 | No |
| 46 | Sparc | 11998 | 0.070 | -0.0335 | No |
| 47 | Lta4h | 12083 | 0.066 | -0.0346 | No |
| 48 | Sirt2 | 12512 | 0.050 | -0.0488 | No |
| 49 | Cpn1 | 12682 | 0.043 | -0.0537 | No |
| 50 | Ctsk | 13632 | 0.012 | -0.0879 | No |
| 51 | Ctsb | 13657 | 0.011 | -0.0885 | No |
| 52 | Ctsl | 13713 | 0.010 | -0.0902 | No |
| 53 | Crip2 | 14418 | -0.001 | -0.1158 | No |
| 54 | Arf4 | 14502 | -0.004 | -0.1187 | No |
| 55 | Itgb3 | 14597 | -0.007 | -0.1219 | No |
| 56 | Klk8 | 14615 | -0.008 | -0.1223 | No |
| 57 | F10 | 14629 | -0.009 | -0.1225 | No |
| 58 | Hpn | 14855 | -0.017 | -0.1302 | No |
| 59 | C8b | 15037 | -0.024 | -0.1361 | No |
| 60 | Gp9 | 15375 | -0.036 | -0.1474 | No |
| 61 | Ctse | 15435 | -0.038 | -0.1485 | No |
| 62 | Olr1 | 15575 | -0.043 | -0.1523 | No |
| 63 | Bmp1 | 16201 | -0.064 | -0.1732 | No |
| 64 | A2m | 16513 | -0.076 | -0.1824 | No |
| 65 | Rabif | 16576 | -0.077 | -0.1825 | No |
| 66 | Comp | 17195 | -0.100 | -0.2021 | No |
| 67 | C1qa | 17472 | -0.110 | -0.2091 | No |
| 68 | Mep1a | 18104 | -0.131 | -0.2283 | No |
| 69 | Ctso | 18217 | -0.136 | -0.2286 | No |
| 70 | F2rl2 | 18620 | -0.151 | -0.2389 | No |
| 71 | Klf7 | 18936 | -0.161 | -0.2458 | No |
| 72 | Wdr1 | 19081 | -0.166 | -0.2464 | No |
| 73 | Trf | 19118 | -0.168 | -0.2429 | No |
| 74 | Tmprss6 | 19120 | -0.168 | -0.2382 | No |
| 75 | Gsn | 19173 | -0.170 | -0.2353 | No |
| 76 | Serpine1 | 19179 | -0.170 | -0.2307 | No |
| 77 | C8a | 19419 | -0.179 | -0.2343 | No |
| 78 | Adam9 | 19503 | -0.182 | -0.2322 | No |
| 79 | Mmp11 | 19527 | -0.183 | -0.2279 | No |
| 80 | Hmgcs2 | 20406 | -0.214 | -0.2538 | No |
| 81 | Gp1ba | 20776 | -0.223 | -0.2609 | No |
| 82 | Mmp14 | 20931 | -0.229 | -0.2600 | No |
| 83 | Rac1 | 21195 | -0.241 | -0.2628 | No |
| 84 | Timp3 | 21201 | -0.241 | -0.2561 | No |
| 85 | F9 | 21618 | -0.260 | -0.2639 | No |
| 86 | Mmp15 | 22034 | -0.275 | -0.2713 | No |
| 87 | Htra1 | 22349 | -0.289 | -0.2745 | Yes |
| 88 | Rgn | 22360 | -0.289 | -0.2667 | Yes |
| 89 | C1s1 | 22636 | -0.300 | -0.2682 | Yes |
| 90 | C9 | 22947 | -0.314 | -0.2706 | Yes |
| 91 | Lamp2 | 22964 | -0.315 | -0.2623 | Yes |
| 92 | Sh2b2 | 23008 | -0.317 | -0.2549 | Yes |
| 93 | F3 | 23010 | -0.317 | -0.2460 | Yes |
| 94 | Iscu | 23090 | -0.321 | -0.2398 | Yes |
| 95 | Fbn1 | 23703 | -0.355 | -0.2520 | Yes |
| 96 | Plek | 23870 | -0.364 | -0.2478 | Yes |
| 97 | Msrb2 | 24006 | -0.372 | -0.2422 | Yes |
| 98 | F11 | 24066 | -0.375 | -0.2338 | Yes |
| 99 | Usp11 | 24148 | -0.379 | -0.2260 | Yes |
| 100 | Mst1 | 24379 | -0.394 | -0.2232 | Yes |
| 101 | Fyn | 24520 | -0.404 | -0.2169 | Yes |
| 102 | Dpp4 | 25051 | -0.436 | -0.2239 | Yes |
| 103 | Klkb1 | 25071 | -0.438 | -0.2122 | Yes |
| 104 | Anxa1 | 25290 | -0.456 | -0.2072 | Yes |
| 105 | Csrp1 | 25387 | -0.464 | -0.1976 | Yes |
| 106 | S100a13 | 25603 | -0.485 | -0.1917 | Yes |
| 107 | Maff | 25623 | -0.486 | -0.1787 | Yes |
| 108 | Furin | 25646 | -0.488 | -0.1657 | Yes |
| 109 | Cpq | 25676 | -0.491 | -0.1528 | Yes |
| 110 | Gnb2 | 25961 | -0.522 | -0.1484 | Yes |
| 111 | Pf4 | 26336 | -0.567 | -0.1460 | Yes |
| 112 | Hnf4a | 26404 | -0.578 | -0.1321 | Yes |
| 113 | Itga2 | 26489 | -0.591 | -0.1185 | Yes |
| 114 | S100a1 | 26559 | -0.600 | -0.1040 | Yes |
| 115 | Thbd | 26650 | -0.614 | -0.0900 | Yes |
| 116 | Gng12 | 26985 | -0.677 | -0.0830 | Yes |
| 117 | Rapgef3 | 27019 | -0.684 | -0.0648 | Yes |
| 118 | C2 | 27103 | -0.708 | -0.0479 | Yes |
| 119 | Pecam1 | 27109 | -0.711 | -0.0279 | Yes |
| 120 | Cd9 | 27120 | -0.714 | -0.0081 | Yes |
| 121 | Capn2 | 27502 | -0.907 | 0.0036 | Yes |