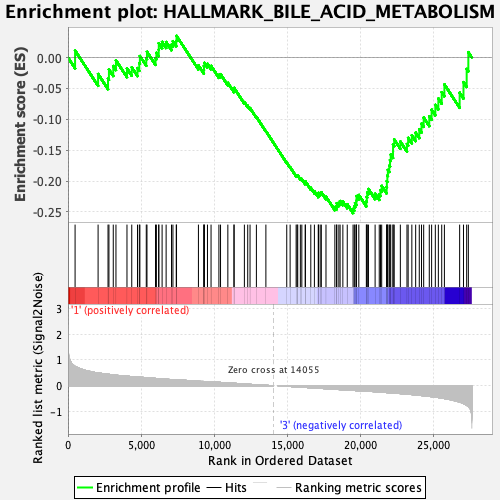
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
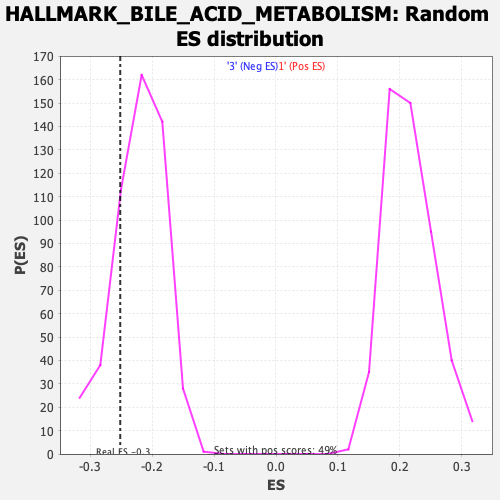
| Enrichment Score (ES) | -0.25184488 |
| Normalized Enrichment Score (NES) | -1.1439861 |
| Nominal p-value | 0.20472442 |
| FDR q-value | 0.6954206 |
| FWER p-Value | 0.95 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca4 | 484 | 0.746 | 0.0113 | No |
| 2 | Ephx2 | 2057 | 0.497 | -0.0267 | No |
| 3 | Gnpat | 2736 | 0.444 | -0.0342 | No |
| 4 | Slco1a4 | 2798 | 0.441 | -0.0193 | No |
| 5 | Aldh1a1 | 3094 | 0.421 | -0.0138 | No |
| 6 | Apoa1 | 3278 | 0.409 | -0.0046 | No |
| 7 | Pfkm | 4031 | 0.370 | -0.0176 | No |
| 8 | Pex6 | 4358 | 0.352 | -0.0159 | No |
| 9 | Idi1 | 4750 | 0.336 | -0.0171 | No |
| 10 | Abca3 | 4874 | 0.330 | -0.0088 | No |
| 11 | Abca5 | 4913 | 0.327 | 0.0024 | No |
| 12 | Acsl5 | 5350 | 0.316 | -0.0012 | No |
| 13 | Slc29a1 | 5398 | 0.313 | 0.0092 | No |
| 14 | Nr1h4 | 5982 | 0.283 | -0.0010 | No |
| 15 | Abcd2 | 6042 | 0.280 | 0.0076 | No |
| 16 | Paox | 6201 | 0.277 | 0.0126 | No |
| 17 | Dhcr24 | 6207 | 0.276 | 0.0231 | No |
| 18 | Isoc1 | 6432 | 0.267 | 0.0253 | No |
| 19 | Gc | 6707 | 0.252 | 0.0251 | No |
| 20 | Abcd3 | 7075 | 0.236 | 0.0209 | No |
| 21 | Retsat | 7172 | 0.233 | 0.0264 | No |
| 22 | Slc23a1 | 7399 | 0.227 | 0.0269 | No |
| 23 | Gclm | 7418 | 0.226 | 0.0350 | No |
| 24 | Pxmp2 | 8915 | 0.172 | -0.0127 | No |
| 25 | Nedd4 | 9285 | 0.163 | -0.0198 | No |
| 26 | Aldh9a1 | 9292 | 0.163 | -0.0137 | No |
| 27 | Slc35b2 | 9324 | 0.162 | -0.0086 | No |
| 28 | Abca6 | 9533 | 0.154 | -0.0102 | No |
| 29 | Scp2 | 9778 | 0.145 | -0.0135 | No |
| 30 | Slc27a2 | 10315 | 0.131 | -0.0279 | No |
| 31 | Abcg4 | 10426 | 0.127 | -0.0270 | No |
| 32 | Nr0b2 | 10926 | 0.109 | -0.0410 | No |
| 33 | Nudt12 | 11347 | 0.093 | -0.0526 | No |
| 34 | Hao1 | 11351 | 0.093 | -0.0491 | No |
| 35 | Aldh8a1 | 12056 | 0.068 | -0.0721 | No |
| 36 | Acsl1 | 12285 | 0.059 | -0.0781 | No |
| 37 | Cyp7b1 | 12443 | 0.053 | -0.0818 | No |
| 38 | Bmp6 | 12877 | 0.036 | -0.0962 | No |
| 39 | Fads2 | 13526 | 0.016 | -0.1191 | No |
| 40 | Sult1b1 | 14952 | -0.020 | -0.1702 | No |
| 41 | Akr1d1 | 15183 | -0.028 | -0.1774 | No |
| 42 | Fads1 | 15594 | -0.043 | -0.1907 | No |
| 43 | Npc1 | 15660 | -0.046 | -0.1912 | No |
| 44 | Abcg8 | 15703 | -0.048 | -0.1909 | No |
| 45 | Rxra | 15882 | -0.054 | -0.1953 | No |
| 46 | Amacr | 15983 | -0.057 | -0.1967 | No |
| 47 | Cyp46a1 | 16214 | -0.065 | -0.2026 | No |
| 48 | Pex19 | 16229 | -0.065 | -0.2006 | No |
| 49 | Slc23a2 | 16596 | -0.078 | -0.2109 | No |
| 50 | Pex16 | 16842 | -0.087 | -0.2164 | No |
| 51 | Cyp39a1 | 17106 | -0.097 | -0.2222 | No |
| 52 | Dio1 | 17122 | -0.097 | -0.2190 | No |
| 53 | Pnpla8 | 17236 | -0.101 | -0.2192 | No |
| 54 | Pex13 | 17316 | -0.104 | -0.2181 | No |
| 55 | Gnmt | 17634 | -0.115 | -0.2251 | No |
| 56 | Klf1 | 18241 | -0.137 | -0.2419 | No |
| 57 | Sult2b1 | 18358 | -0.142 | -0.2406 | No |
| 58 | Lonp2 | 18375 | -0.142 | -0.2357 | No |
| 59 | Abca8b | 18515 | -0.147 | -0.2351 | No |
| 60 | Pex7 | 18599 | -0.150 | -0.2323 | No |
| 61 | Slc27a5 | 18788 | -0.158 | -0.2330 | No |
| 62 | Pecr | 19088 | -0.166 | -0.2374 | No |
| 63 | Optn | 19485 | -0.181 | -0.2448 | Yes |
| 64 | Hsd17b4 | 19570 | -0.184 | -0.2408 | Yes |
| 65 | Abcd1 | 19624 | -0.186 | -0.2355 | Yes |
| 66 | Mlycd | 19717 | -0.190 | -0.2315 | Yes |
| 67 | Cat | 19721 | -0.190 | -0.2243 | Yes |
| 68 | Rbp1 | 19878 | -0.195 | -0.2224 | Yes |
| 69 | Crot | 20393 | -0.214 | -0.2328 | Yes |
| 70 | Pex12 | 20423 | -0.215 | -0.2256 | Yes |
| 71 | Cyp27a1 | 20455 | -0.217 | -0.2183 | Yes |
| 72 | Pex1 | 20537 | -0.220 | -0.2128 | Yes |
| 73 | Tfcp2l1 | 20992 | -0.232 | -0.2203 | Yes |
| 74 | Idh2 | 21275 | -0.244 | -0.2211 | Yes |
| 75 | Aqp9 | 21362 | -0.249 | -0.2146 | Yes |
| 76 | Bbox1 | 21442 | -0.252 | -0.2077 | Yes |
| 77 | Fdxr | 21779 | -0.264 | -0.2097 | Yes |
| 78 | Efhc1 | 21795 | -0.265 | -0.2000 | Yes |
| 79 | Atxn1 | 21829 | -0.267 | -0.1909 | Yes |
| 80 | Sod1 | 21865 | -0.268 | -0.1818 | Yes |
| 81 | Lipe | 21963 | -0.272 | -0.1748 | Yes |
| 82 | Pex26 | 22012 | -0.274 | -0.1660 | Yes |
| 83 | Pex11a | 22060 | -0.276 | -0.1570 | Yes |
| 84 | Bcar3 | 22217 | -0.283 | -0.1517 | Yes |
| 85 | Gstk1 | 22219 | -0.284 | -0.1408 | Yes |
| 86 | Nr1i2 | 22298 | -0.287 | -0.1325 | Yes |
| 87 | Ar | 22717 | -0.304 | -0.1360 | Yes |
| 88 | Abca9 | 23170 | -0.325 | -0.1398 | Yes |
| 89 | Hacl1 | 23250 | -0.329 | -0.1300 | Yes |
| 90 | Idh1 | 23503 | -0.342 | -0.1259 | Yes |
| 91 | Pipox | 23764 | -0.358 | -0.1215 | Yes |
| 92 | Rxrg | 24011 | -0.372 | -0.1161 | Yes |
| 93 | Pex11g | 24164 | -0.380 | -0.1069 | Yes |
| 94 | Dio2 | 24312 | -0.390 | -0.0972 | Yes |
| 95 | Nr3c2 | 24695 | -0.413 | -0.0951 | Yes |
| 96 | Soat2 | 24855 | -0.424 | -0.0845 | Yes |
| 97 | Prdx5 | 25108 | -0.441 | -0.0766 | Yes |
| 98 | Abca2 | 25315 | -0.458 | -0.0664 | Yes |
| 99 | Lck | 25545 | -0.480 | -0.0561 | Yes |
| 100 | Abca1 | 25728 | -0.497 | -0.0435 | Yes |
| 101 | Phyh | 26771 | -0.632 | -0.0570 | Yes |
| 102 | Hsd17b11 | 27033 | -0.688 | -0.0399 | Yes |
| 103 | Slc22a18 | 27251 | -0.758 | -0.0185 | Yes |
| 104 | Hsd3b7 | 27365 | -0.806 | 0.0086 | Yes |