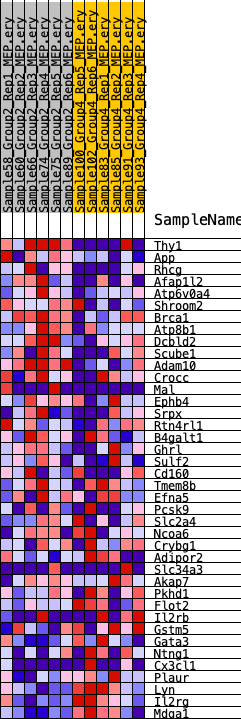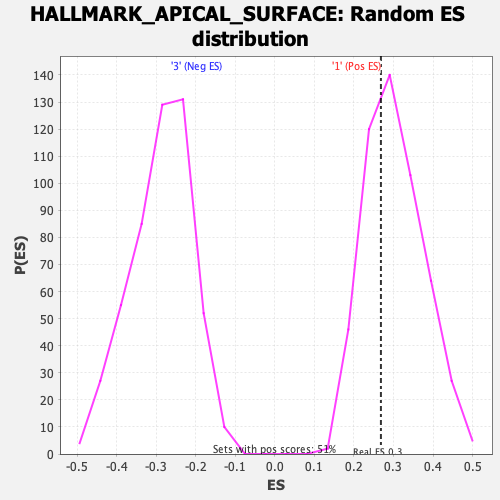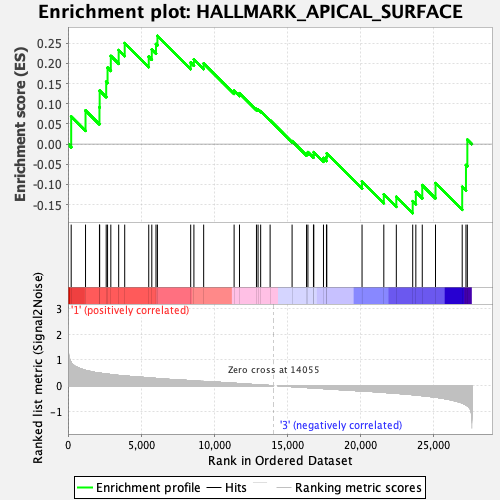
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group4.MEP.ery_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_SURFACE |
| Enrichment Score (ES) | 0.2682563 |
| Normalized Enrichment Score (NES) | 0.8901377 |
| Nominal p-value | 0.6311637 |
| FDR q-value | 0.78043365 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thy1 | 218 | 0.892 | 0.0684 | Yes |
| 2 | App | 1198 | 0.592 | 0.0835 | Yes |
| 3 | Rhcg | 2150 | 0.493 | 0.0911 | Yes |
| 4 | Afap1l2 | 2165 | 0.491 | 0.1326 | Yes |
| 5 | Atp6v0a4 | 2611 | 0.448 | 0.1548 | Yes |
| 6 | Shroom2 | 2704 | 0.445 | 0.1894 | Yes |
| 7 | Brca1 | 2927 | 0.433 | 0.2184 | Yes |
| 8 | Atp8b1 | 3464 | 0.398 | 0.2330 | Yes |
| 9 | Dcbld2 | 3876 | 0.377 | 0.2503 | Yes |
| 10 | Scube1 | 5519 | 0.305 | 0.2168 | Yes |
| 11 | Adam10 | 5728 | 0.295 | 0.2344 | Yes |
| 12 | Crocc | 6015 | 0.281 | 0.2481 | Yes |
| 13 | Mal | 6113 | 0.277 | 0.2683 | Yes |
| 14 | Ephb4 | 8386 | 0.193 | 0.2023 | No |
| 15 | Srpx | 8606 | 0.185 | 0.2101 | No |
| 16 | Rtn4rl1 | 9273 | 0.164 | 0.2000 | No |
| 17 | B4galt1 | 11354 | 0.093 | 0.1325 | No |
| 18 | Ghrl | 11722 | 0.080 | 0.1260 | No |
| 19 | Sulf2 | 12882 | 0.036 | 0.0870 | No |
| 20 | Cd160 | 12991 | 0.032 | 0.0859 | No |
| 21 | Tmem8b | 13169 | 0.026 | 0.0817 | No |
| 22 | Efna5 | 13815 | 0.006 | 0.0588 | No |
| 23 | Pcsk9 | 15317 | -0.033 | 0.0072 | No |
| 24 | Slc2a4 | 16305 | -0.068 | -0.0228 | No |
| 25 | Ncoa6 | 16399 | -0.072 | -0.0200 | No |
| 26 | Crybg1 | 16778 | -0.085 | -0.0265 | No |
| 27 | Adipor2 | 16802 | -0.086 | -0.0200 | No |
| 28 | Slc34a3 | 17463 | -0.110 | -0.0346 | No |
| 29 | Akap7 | 17667 | -0.116 | -0.0320 | No |
| 30 | Pkhd1 | 17690 | -0.117 | -0.0228 | No |
| 31 | Flot2 | 20099 | -0.204 | -0.0928 | No |
| 32 | Il2rb | 21587 | -0.259 | -0.1246 | No |
| 33 | Gstm5 | 22442 | -0.292 | -0.1306 | No |
| 34 | Gata3 | 23562 | -0.346 | -0.1416 | No |
| 35 | Ntng1 | 23776 | -0.359 | -0.1187 | No |
| 36 | Cx3cl1 | 24215 | -0.384 | -0.1017 | No |
| 37 | Plaur | 25120 | -0.442 | -0.0968 | No |
| 38 | Lyn | 26945 | -0.667 | -0.1059 | No |
| 39 | Il2rg | 27203 | -0.742 | -0.0518 | No |
| 40 | Mdga1 | 27297 | -0.775 | 0.0111 | No |