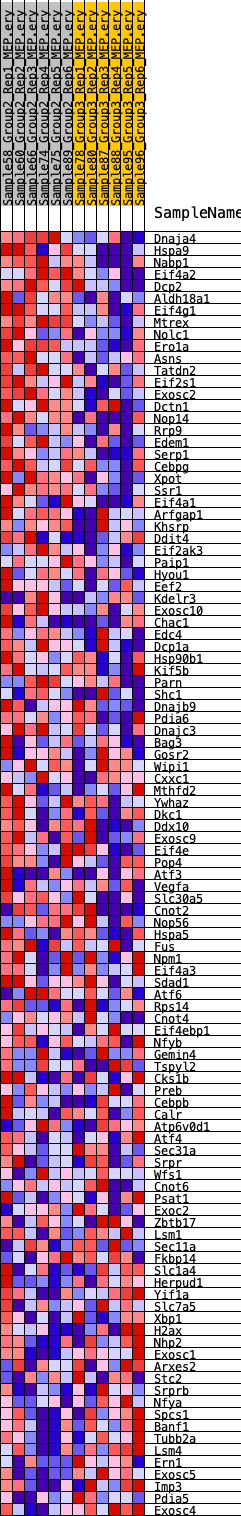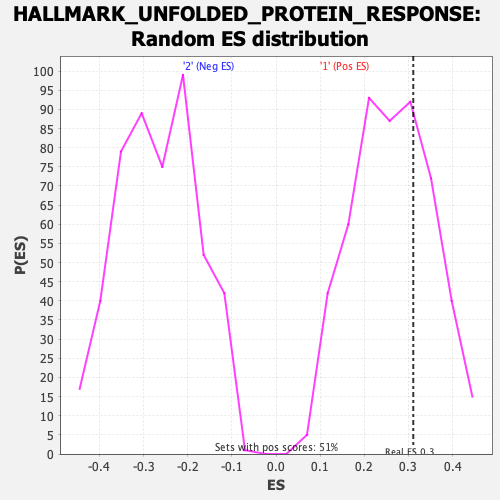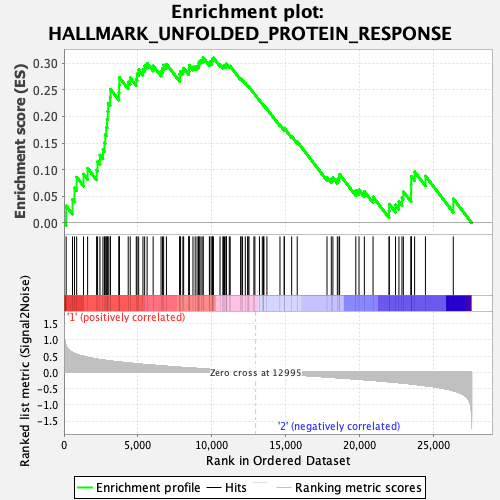
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.31097037 |
| Normalized Enrichment Score (NES) | 1.1841053 |
| Nominal p-value | 0.3241107 |
| FDR q-value | 0.56182426 |
| FWER p-Value | 0.948 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dnaja4 | 146 | 0.808 | 0.0317 | Yes |
| 2 | Hspa9 | 576 | 0.609 | 0.0440 | Yes |
| 3 | Nabp1 | 711 | 0.578 | 0.0657 | Yes |
| 4 | Eif4a2 | 858 | 0.555 | 0.0858 | Yes |
| 5 | Dcp2 | 1321 | 0.494 | 0.0917 | Yes |
| 6 | Aldh18a1 | 1600 | 0.458 | 0.1025 | Yes |
| 7 | Eif4g1 | 2207 | 0.407 | 0.0992 | Yes |
| 8 | Mtrex | 2273 | 0.403 | 0.1153 | Yes |
| 9 | Nolc1 | 2434 | 0.392 | 0.1274 | Yes |
| 10 | Ero1a | 2638 | 0.380 | 0.1374 | Yes |
| 11 | Asns | 2740 | 0.373 | 0.1509 | Yes |
| 12 | Tatdn2 | 2794 | 0.370 | 0.1659 | Yes |
| 13 | Eif2s1 | 2865 | 0.365 | 0.1801 | Yes |
| 14 | Exosc2 | 2919 | 0.362 | 0.1947 | Yes |
| 15 | Dctn1 | 2963 | 0.359 | 0.2096 | Yes |
| 16 | Nop14 | 2991 | 0.357 | 0.2250 | Yes |
| 17 | Rrp9 | 3123 | 0.348 | 0.2362 | Yes |
| 18 | Edem1 | 3144 | 0.347 | 0.2513 | Yes |
| 19 | Serp1 | 3721 | 0.321 | 0.2451 | Yes |
| 20 | Cebpg | 3734 | 0.320 | 0.2593 | Yes |
| 21 | Xpot | 3750 | 0.319 | 0.2734 | Yes |
| 22 | Ssr1 | 4348 | 0.288 | 0.2648 | Yes |
| 23 | Eif4a1 | 4484 | 0.281 | 0.2728 | Yes |
| 24 | Arfgap1 | 4893 | 0.265 | 0.2701 | Yes |
| 25 | Khsrp | 4947 | 0.262 | 0.2802 | Yes |
| 26 | Ddit4 | 5056 | 0.257 | 0.2880 | Yes |
| 27 | Eif2ak3 | 5347 | 0.244 | 0.2886 | Yes |
| 28 | Paip1 | 5461 | 0.238 | 0.2955 | Yes |
| 29 | Hyou1 | 5628 | 0.232 | 0.3001 | Yes |
| 30 | Eef2 | 6035 | 0.219 | 0.2954 | Yes |
| 31 | Kdelr3 | 6572 | 0.201 | 0.2851 | Yes |
| 32 | Exosc10 | 6668 | 0.196 | 0.2906 | Yes |
| 33 | Chac1 | 6747 | 0.193 | 0.2966 | Yes |
| 34 | Edc4 | 6936 | 0.186 | 0.2983 | Yes |
| 35 | Dcp1a | 7834 | 0.156 | 0.2728 | Yes |
| 36 | Hsp90b1 | 7841 | 0.156 | 0.2797 | Yes |
| 37 | Kif5b | 7894 | 0.154 | 0.2849 | Yes |
| 38 | Parn | 8051 | 0.148 | 0.2860 | Yes |
| 39 | Shc1 | 8102 | 0.146 | 0.2909 | Yes |
| 40 | Dnajb9 | 8442 | 0.139 | 0.2849 | Yes |
| 41 | Pdia6 | 8471 | 0.138 | 0.2903 | Yes |
| 42 | Dnajc3 | 8482 | 0.138 | 0.2962 | Yes |
| 43 | Bag3 | 8734 | 0.130 | 0.2930 | Yes |
| 44 | Gosr2 | 8883 | 0.125 | 0.2934 | Yes |
| 45 | Wipi1 | 9012 | 0.121 | 0.2943 | Yes |
| 46 | Cxxc1 | 9115 | 0.118 | 0.2960 | Yes |
| 47 | Mthfd2 | 9132 | 0.117 | 0.3008 | Yes |
| 48 | Ywhaz | 9192 | 0.115 | 0.3039 | Yes |
| 49 | Dkc1 | 9281 | 0.113 | 0.3059 | Yes |
| 50 | Ddx10 | 9415 | 0.108 | 0.3060 | Yes |
| 51 | Exosc9 | 9416 | 0.108 | 0.3110 | Yes |
| 52 | Eif4e | 9846 | 0.094 | 0.2997 | No |
| 53 | Pop4 | 9883 | 0.093 | 0.3026 | No |
| 54 | Atf3 | 10021 | 0.089 | 0.3017 | No |
| 55 | Vegfa | 10030 | 0.089 | 0.3055 | No |
| 56 | Slc30a5 | 10053 | 0.088 | 0.3088 | No |
| 57 | Cnot2 | 10120 | 0.086 | 0.3103 | No |
| 58 | Nop56 | 10565 | 0.073 | 0.2975 | No |
| 59 | Hspa5 | 10760 | 0.066 | 0.2935 | No |
| 60 | Fus | 10796 | 0.065 | 0.2952 | No |
| 61 | Npm1 | 10870 | 0.063 | 0.2954 | No |
| 62 | Eif4a3 | 10970 | 0.060 | 0.2946 | No |
| 63 | Sdad1 | 10992 | 0.059 | 0.2965 | No |
| 64 | Atf6 | 10995 | 0.059 | 0.2992 | No |
| 65 | Rps14 | 11193 | 0.053 | 0.2944 | No |
| 66 | Cnot4 | 11253 | 0.051 | 0.2946 | No |
| 67 | Eif4ebp1 | 11961 | 0.029 | 0.2702 | No |
| 68 | Nfyb | 11991 | 0.029 | 0.2705 | No |
| 69 | Gemin4 | 12091 | 0.025 | 0.2681 | No |
| 70 | Tspyl2 | 12277 | 0.020 | 0.2623 | No |
| 71 | Cks1b | 12434 | 0.016 | 0.2573 | No |
| 72 | Preb | 12466 | 0.015 | 0.2569 | No |
| 73 | Cebpb | 12527 | 0.014 | 0.2554 | No |
| 74 | Calr | 12859 | 0.004 | 0.2435 | No |
| 75 | Atp6v0d1 | 12926 | 0.002 | 0.2412 | No |
| 76 | Atf4 | 13236 | -0.002 | 0.2301 | No |
| 77 | Sec31a | 13428 | -0.009 | 0.2235 | No |
| 78 | Srpr | 13508 | -0.011 | 0.2211 | No |
| 79 | Wfs1 | 13539 | -0.011 | 0.2206 | No |
| 80 | Cnot6 | 13740 | -0.017 | 0.2141 | No |
| 81 | Psat1 | 14624 | -0.042 | 0.1839 | No |
| 82 | Exoc2 | 14912 | -0.051 | 0.1758 | No |
| 83 | Zbtb17 | 14917 | -0.051 | 0.1780 | No |
| 84 | Lsm1 | 15416 | -0.065 | 0.1628 | No |
| 85 | Sec11a | 15792 | -0.077 | 0.1527 | No |
| 86 | Fkbp14 | 17809 | -0.139 | 0.0857 | No |
| 87 | Slc1a4 | 18099 | -0.148 | 0.0820 | No |
| 88 | Herpud1 | 18209 | -0.150 | 0.0849 | No |
| 89 | Yif1a | 18508 | -0.160 | 0.0814 | No |
| 90 | Slc7a5 | 18623 | -0.164 | 0.0848 | No |
| 91 | Xbp1 | 18662 | -0.166 | 0.0910 | No |
| 92 | H2ax | 19759 | -0.201 | 0.0604 | No |
| 93 | Nhp2 | 19977 | -0.209 | 0.0621 | No |
| 94 | Exosc1 | 20338 | -0.220 | 0.0591 | No |
| 95 | Arxes2 | 20927 | -0.241 | 0.0487 | No |
| 96 | Stc2 | 22011 | -0.286 | 0.0225 | No |
| 97 | Srprb | 22026 | -0.287 | 0.0351 | No |
| 98 | Nfya | 22450 | -0.304 | 0.0336 | No |
| 99 | Spcs1 | 22680 | -0.313 | 0.0397 | No |
| 100 | Banf1 | 22885 | -0.322 | 0.0470 | No |
| 101 | Tubb2a | 22979 | -0.325 | 0.0585 | No |
| 102 | Lsm4 | 23505 | -0.352 | 0.0556 | No |
| 103 | Ern1 | 23506 | -0.352 | 0.0717 | No |
| 104 | Exosc5 | 23510 | -0.352 | 0.0877 | No |
| 105 | Imp3 | 23742 | -0.364 | 0.0960 | No |
| 106 | Pdia5 | 24474 | -0.406 | 0.0880 | No |
| 107 | Exosc4 | 26360 | -0.561 | 0.0452 | No |