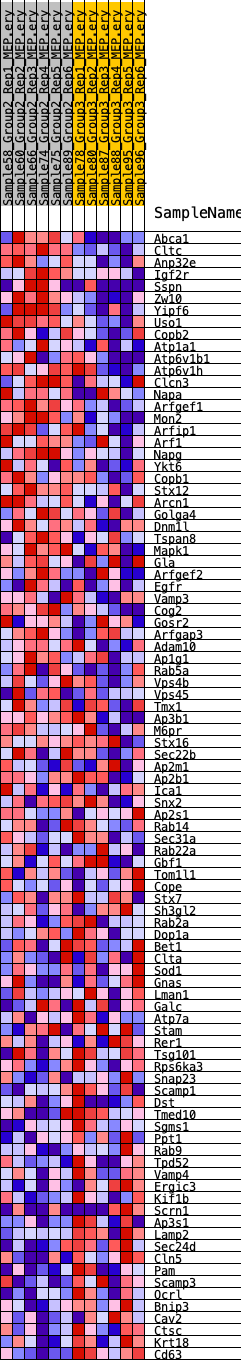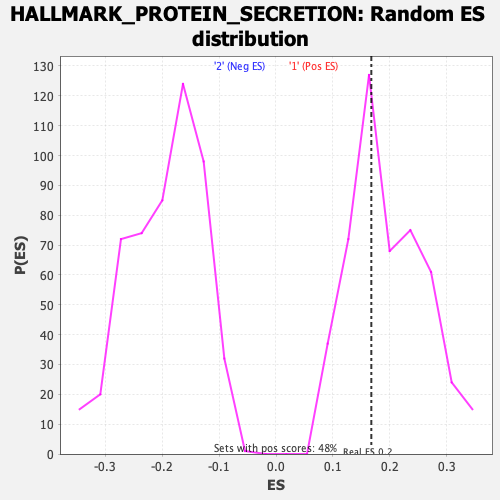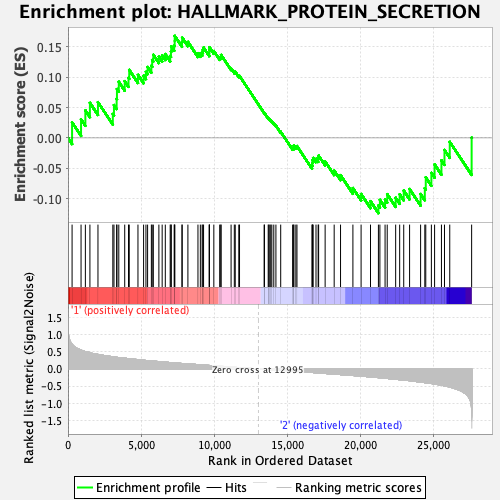
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.16772105 |
| Normalized Enrichment Score (NES) | 0.8564237 |
| Nominal p-value | 0.611691 |
| FDR q-value | 0.9845632 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 279 | 0.714 | 0.0255 | Yes |
| 2 | Cltc | 892 | 0.547 | 0.0306 | Yes |
| 3 | Anp32e | 1187 | 0.506 | 0.0452 | Yes |
| 4 | Igf2r | 1498 | 0.474 | 0.0576 | Yes |
| 5 | Sspn | 2048 | 0.422 | 0.0588 | Yes |
| 6 | Zw10 | 3064 | 0.353 | 0.0395 | Yes |
| 7 | Yipf6 | 3142 | 0.347 | 0.0540 | Yes |
| 8 | Uso1 | 3324 | 0.338 | 0.0643 | Yes |
| 9 | Copb2 | 3346 | 0.337 | 0.0804 | Yes |
| 10 | Atp1a1 | 3472 | 0.329 | 0.0923 | Yes |
| 11 | Atp6v1b1 | 3877 | 0.314 | 0.0933 | Yes |
| 12 | Atp6v1h | 4135 | 0.299 | 0.0988 | Yes |
| 13 | Clcn3 | 4186 | 0.296 | 0.1118 | Yes |
| 14 | Napa | 4782 | 0.270 | 0.1037 | Yes |
| 15 | Arfgef1 | 5167 | 0.252 | 0.1023 | Yes |
| 16 | Mon2 | 5328 | 0.244 | 0.1087 | Yes |
| 17 | Arfip1 | 5441 | 0.239 | 0.1166 | Yes |
| 18 | Arf1 | 5698 | 0.231 | 0.1188 | Yes |
| 19 | Napg | 5755 | 0.229 | 0.1282 | Yes |
| 20 | Ykt6 | 5832 | 0.226 | 0.1367 | Yes |
| 21 | Copb1 | 6212 | 0.213 | 0.1336 | Yes |
| 22 | Stx12 | 6436 | 0.206 | 0.1357 | Yes |
| 23 | Arcn1 | 6651 | 0.198 | 0.1378 | Yes |
| 24 | Golga4 | 6985 | 0.184 | 0.1349 | Yes |
| 25 | Dnm1l | 7038 | 0.182 | 0.1421 | Yes |
| 26 | Tspan8 | 7056 | 0.181 | 0.1505 | Yes |
| 27 | Mapk1 | 7258 | 0.173 | 0.1518 | Yes |
| 28 | Gla | 7292 | 0.172 | 0.1592 | Yes |
| 29 | Arfgef2 | 7294 | 0.172 | 0.1677 | Yes |
| 30 | Egfr | 7790 | 0.157 | 0.1576 | No |
| 31 | Vamp3 | 7804 | 0.157 | 0.1649 | No |
| 32 | Cog2 | 8196 | 0.143 | 0.1579 | No |
| 33 | Gosr2 | 8883 | 0.125 | 0.1392 | No |
| 34 | Arfgap3 | 9047 | 0.120 | 0.1392 | No |
| 35 | Adam10 | 9168 | 0.116 | 0.1407 | No |
| 36 | Ap1g1 | 9218 | 0.115 | 0.1446 | No |
| 37 | Rab5a | 9266 | 0.113 | 0.1486 | No |
| 38 | Vps4b | 9657 | 0.100 | 0.1394 | No |
| 39 | Vps45 | 9658 | 0.100 | 0.1444 | No |
| 40 | Tmx1 | 9669 | 0.100 | 0.1491 | No |
| 41 | Ap3b1 | 9968 | 0.091 | 0.1428 | No |
| 42 | M6pr | 10369 | 0.079 | 0.1321 | No |
| 43 | Stx16 | 10444 | 0.077 | 0.1333 | No |
| 44 | Sec22b | 10464 | 0.076 | 0.1364 | No |
| 45 | Ap2m1 | 11147 | 0.054 | 0.1143 | No |
| 46 | Ap2b1 | 11368 | 0.048 | 0.1087 | No |
| 47 | Ica1 | 11431 | 0.045 | 0.1087 | No |
| 48 | Snx2 | 11675 | 0.038 | 0.1018 | No |
| 49 | Ap2s1 | 11724 | 0.037 | 0.1019 | No |
| 50 | Rab14 | 13408 | -0.008 | 0.0411 | No |
| 51 | Sec31a | 13428 | -0.009 | 0.0408 | No |
| 52 | Rab22a | 13685 | -0.015 | 0.0323 | No |
| 53 | Gbf1 | 13728 | -0.017 | 0.0316 | No |
| 54 | Tom1l1 | 13796 | -0.019 | 0.0301 | No |
| 55 | Cope | 13862 | -0.020 | 0.0287 | No |
| 56 | Stx7 | 13933 | -0.022 | 0.0273 | No |
| 57 | Sh3gl2 | 14066 | -0.026 | 0.0238 | No |
| 58 | Rab2a | 14212 | -0.030 | 0.0200 | No |
| 59 | Dop1a | 14539 | -0.039 | 0.0101 | No |
| 60 | Bet1 | 15353 | -0.063 | -0.0163 | No |
| 61 | Clta | 15431 | -0.065 | -0.0159 | No |
| 62 | Sod1 | 15433 | -0.065 | -0.0126 | No |
| 63 | Gnas | 15575 | -0.070 | -0.0143 | No |
| 64 | Lman1 | 15650 | -0.072 | -0.0133 | No |
| 65 | Galc | 16677 | -0.102 | -0.0455 | No |
| 66 | Atp7a | 16705 | -0.103 | -0.0414 | No |
| 67 | Stam | 16712 | -0.103 | -0.0365 | No |
| 68 | Rer1 | 16759 | -0.105 | -0.0329 | No |
| 69 | Tsg101 | 16952 | -0.112 | -0.0343 | No |
| 70 | Rps6ka3 | 17098 | -0.116 | -0.0338 | No |
| 71 | Snap23 | 17131 | -0.117 | -0.0291 | No |
| 72 | Scamp1 | 17576 | -0.131 | -0.0387 | No |
| 73 | Dst | 18194 | -0.150 | -0.0536 | No |
| 74 | Tmed10 | 18629 | -0.164 | -0.0612 | No |
| 75 | Sgms1 | 19474 | -0.191 | -0.0824 | No |
| 76 | Ppt1 | 20034 | -0.211 | -0.0922 | No |
| 77 | Rab9 | 20677 | -0.230 | -0.1040 | No |
| 78 | Tpd52 | 21218 | -0.253 | -0.1110 | No |
| 79 | Vamp4 | 21319 | -0.257 | -0.1018 | No |
| 80 | Ergic3 | 21673 | -0.271 | -0.1011 | No |
| 81 | Kif1b | 21820 | -0.278 | -0.0925 | No |
| 82 | Scrn1 | 22397 | -0.301 | -0.0984 | No |
| 83 | Ap3s1 | 22671 | -0.312 | -0.0927 | No |
| 84 | Lamp2 | 22948 | -0.324 | -0.0865 | No |
| 85 | Sec24d | 23349 | -0.342 | -0.0840 | No |
| 86 | Cln5 | 24102 | -0.383 | -0.0922 | No |
| 87 | Pam | 24390 | -0.400 | -0.0827 | No |
| 88 | Scamp3 | 24455 | -0.405 | -0.0648 | No |
| 89 | Ocrl | 24838 | -0.424 | -0.0575 | No |
| 90 | Bnip3 | 25058 | -0.439 | -0.0435 | No |
| 91 | Cav2 | 25526 | -0.474 | -0.0368 | No |
| 92 | Ctsc | 25734 | -0.492 | -0.0198 | No |
| 93 | Krt18 | 26094 | -0.527 | -0.0065 | No |
| 94 | Cd63 | 27590 | -1.227 | 0.0004 | No |