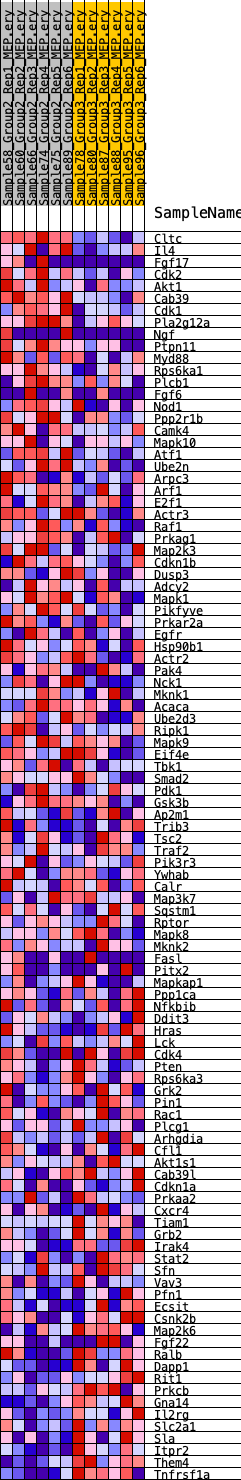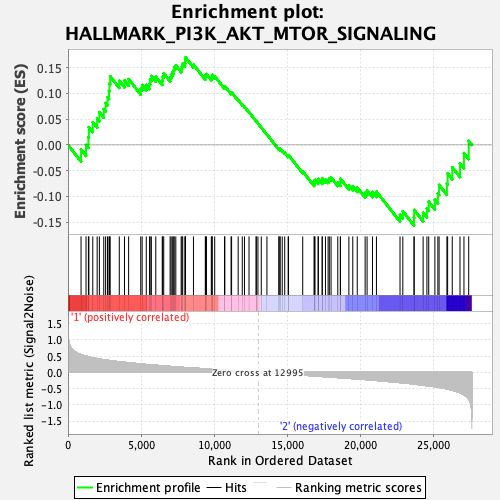
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.17005374 |
| Normalized Enrichment Score (NES) | 0.75587744 |
| Nominal p-value | 0.778 |
| FDR q-value | 0.94822425 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 892 | 0.547 | -0.0090 | Yes |
| 2 | Il4 | 1232 | 0.501 | 0.0001 | Yes |
| 3 | Fgf17 | 1396 | 0.486 | 0.0150 | Yes |
| 4 | Cdk2 | 1428 | 0.483 | 0.0345 | Yes |
| 5 | Akt1 | 1693 | 0.447 | 0.0441 | Yes |
| 6 | Cab39 | 1992 | 0.426 | 0.0515 | Yes |
| 7 | Cdk1 | 2144 | 0.414 | 0.0637 | Yes |
| 8 | Pla2g12a | 2444 | 0.391 | 0.0696 | Yes |
| 9 | Ngf | 2572 | 0.384 | 0.0814 | Yes |
| 10 | Ptpn11 | 2700 | 0.376 | 0.0928 | Yes |
| 11 | Myd88 | 2802 | 0.369 | 0.1050 | Yes |
| 12 | Rps6ka1 | 2835 | 0.367 | 0.1195 | Yes |
| 13 | Plcb1 | 2882 | 0.364 | 0.1334 | Yes |
| 14 | Fgf6 | 3505 | 0.328 | 0.1248 | Yes |
| 15 | Nod1 | 3861 | 0.314 | 0.1254 | Yes |
| 16 | Ppp2r1b | 4145 | 0.298 | 0.1278 | Yes |
| 17 | Camk4 | 4973 | 0.261 | 0.1089 | Yes |
| 18 | Mapk10 | 5068 | 0.256 | 0.1165 | Yes |
| 19 | Atf1 | 5351 | 0.243 | 0.1166 | Yes |
| 20 | Ube2n | 5576 | 0.235 | 0.1185 | Yes |
| 21 | Arpc3 | 5614 | 0.233 | 0.1271 | Yes |
| 22 | Arf1 | 5698 | 0.231 | 0.1340 | Yes |
| 23 | E2f1 | 6001 | 0.221 | 0.1325 | Yes |
| 24 | Actr3 | 6442 | 0.206 | 0.1253 | Yes |
| 25 | Raf1 | 6473 | 0.205 | 0.1330 | Yes |
| 26 | Prkag1 | 6543 | 0.202 | 0.1391 | Yes |
| 27 | Map2k3 | 6980 | 0.184 | 0.1311 | Yes |
| 28 | Cdkn1b | 7048 | 0.181 | 0.1364 | Yes |
| 29 | Dusp3 | 7131 | 0.178 | 0.1411 | Yes |
| 30 | Adcy2 | 7227 | 0.174 | 0.1451 | Yes |
| 31 | Mapk1 | 7258 | 0.173 | 0.1514 | Yes |
| 32 | Pikfyve | 7369 | 0.169 | 0.1546 | Yes |
| 33 | Prkar2a | 7735 | 0.159 | 0.1481 | Yes |
| 34 | Egfr | 7790 | 0.157 | 0.1529 | Yes |
| 35 | Hsp90b1 | 7841 | 0.156 | 0.1577 | Yes |
| 36 | Actr2 | 7993 | 0.150 | 0.1587 | Yes |
| 37 | Pak4 | 8016 | 0.150 | 0.1643 | Yes |
| 38 | Nck1 | 8033 | 0.149 | 0.1701 | Yes |
| 39 | Mknk1 | 8576 | 0.135 | 0.1561 | No |
| 40 | Acaca | 9386 | 0.109 | 0.1314 | No |
| 41 | Ube2d3 | 9398 | 0.109 | 0.1356 | No |
| 42 | Ripk1 | 9465 | 0.106 | 0.1378 | No |
| 43 | Mapk9 | 9795 | 0.096 | 0.1299 | No |
| 44 | Eif4e | 9846 | 0.094 | 0.1321 | No |
| 45 | Tbk1 | 9850 | 0.094 | 0.1361 | No |
| 46 | Smad2 | 10027 | 0.089 | 0.1335 | No |
| 47 | Pdk1 | 10698 | 0.068 | 0.1120 | No |
| 48 | Gsk3b | 10721 | 0.068 | 0.1141 | No |
| 49 | Ap2m1 | 11147 | 0.054 | 0.1010 | No |
| 50 | Trib3 | 11183 | 0.053 | 0.1020 | No |
| 51 | Tsc2 | 11631 | 0.039 | 0.0874 | No |
| 52 | Traf2 | 11911 | 0.031 | 0.0786 | No |
| 53 | Pik3r3 | 12062 | 0.027 | 0.0743 | No |
| 54 | Ywhab | 12373 | 0.017 | 0.0638 | No |
| 55 | Calr | 12859 | 0.004 | 0.0463 | No |
| 56 | Map3k7 | 12867 | 0.004 | 0.0462 | No |
| 57 | Sqstm1 | 12982 | 0.000 | 0.0421 | No |
| 58 | Rptor | 13211 | -0.001 | 0.0338 | No |
| 59 | Mapk8 | 13598 | -0.013 | 0.0204 | No |
| 60 | Mknk2 | 14404 | -0.035 | -0.0074 | No |
| 61 | Fasl | 14480 | -0.037 | -0.0085 | No |
| 62 | Pitx2 | 14497 | -0.038 | -0.0075 | No |
| 63 | Mapkap1 | 14643 | -0.043 | -0.0109 | No |
| 64 | Ppp1ca | 14814 | -0.048 | -0.0151 | No |
| 65 | Nfkbib | 15048 | -0.054 | -0.0212 | No |
| 66 | Ddit3 | 15074 | -0.055 | -0.0198 | No |
| 67 | Hras | 16044 | -0.085 | -0.0514 | No |
| 68 | Lck | 16824 | -0.107 | -0.0751 | No |
| 69 | Cdk4 | 16839 | -0.108 | -0.0710 | No |
| 70 | Pten | 16883 | -0.109 | -0.0679 | No |
| 71 | Rps6ka3 | 17098 | -0.116 | -0.0707 | No |
| 72 | Grk2 | 17109 | -0.116 | -0.0661 | No |
| 73 | Pin1 | 17374 | -0.125 | -0.0704 | No |
| 74 | Rac1 | 17380 | -0.125 | -0.0652 | No |
| 75 | Plcg1 | 17597 | -0.132 | -0.0674 | No |
| 76 | Arhgdia | 17779 | -0.138 | -0.0681 | No |
| 77 | Cfl1 | 17862 | -0.140 | -0.0650 | No |
| 78 | Akt1s1 | 17981 | -0.144 | -0.0632 | No |
| 79 | Cab39l | 18441 | -0.158 | -0.0731 | No |
| 80 | Cdkn1a | 18617 | -0.164 | -0.0725 | No |
| 81 | Prkaa2 | 18622 | -0.164 | -0.0656 | No |
| 82 | Cxcr4 | 19200 | -0.182 | -0.0788 | No |
| 83 | Tiam1 | 19465 | -0.190 | -0.0802 | No |
| 84 | Grb2 | 19771 | -0.201 | -0.0827 | No |
| 85 | Irak4 | 20310 | -0.219 | -0.0929 | No |
| 86 | Stat2 | 20443 | -0.224 | -0.0881 | No |
| 87 | Sfn | 20812 | -0.236 | -0.0914 | No |
| 88 | Vav3 | 21083 | -0.247 | -0.0906 | No |
| 89 | Pfn1 | 22695 | -0.314 | -0.1358 | No |
| 90 | Ecsit | 22886 | -0.322 | -0.1289 | No |
| 91 | Csnk2b | 23652 | -0.359 | -0.1414 | No |
| 92 | Map2k6 | 23662 | -0.360 | -0.1263 | No |
| 93 | Fgf22 | 24274 | -0.392 | -0.1317 | No |
| 94 | Ralb | 24533 | -0.410 | -0.1236 | No |
| 95 | Dapp1 | 24657 | -0.417 | -0.1102 | No |
| 96 | Rit1 | 25078 | -0.440 | -0.1067 | No |
| 97 | Prkcb | 25282 | -0.456 | -0.0945 | No |
| 98 | Gna14 | 25365 | -0.463 | -0.0777 | No |
| 99 | Il2rg | 25900 | -0.507 | -0.0754 | No |
| 100 | Slc2a1 | 25949 | -0.511 | -0.0553 | No |
| 101 | Sla | 26266 | -0.547 | -0.0433 | No |
| 102 | Itpr2 | 26795 | -0.626 | -0.0357 | No |
| 103 | Them4 | 27065 | -0.691 | -0.0159 | No |
| 104 | Tnfrsf1a | 27389 | -0.828 | 0.0077 | No |