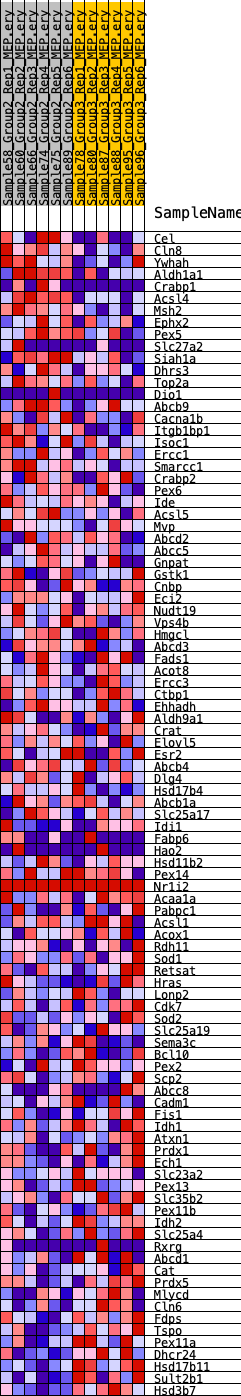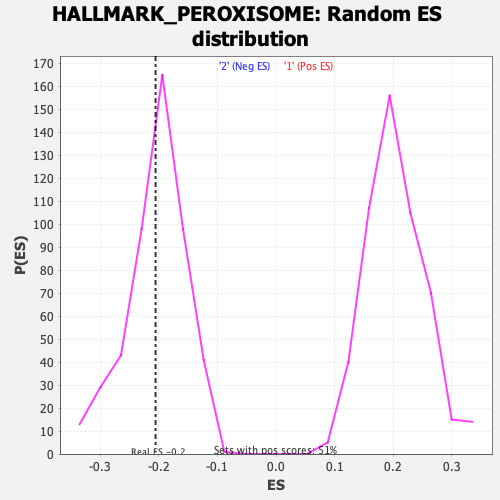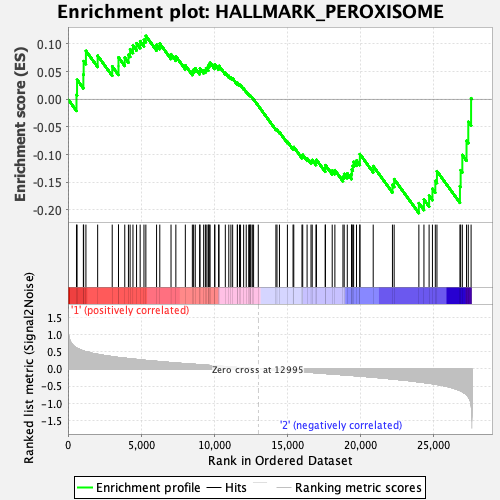
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.20573664 |
| Normalized Enrichment Score (NES) | -1.0031779 |
| Nominal p-value | 0.45491803 |
| FDR q-value | 0.7851659 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cel | 580 | 0.608 | 0.0081 | No |
| 2 | Cln8 | 619 | 0.596 | 0.0352 | No |
| 3 | Ywhah | 1050 | 0.521 | 0.0446 | No |
| 4 | Aldh1a1 | 1070 | 0.518 | 0.0688 | No |
| 5 | Crabp1 | 1225 | 0.503 | 0.0873 | No |
| 6 | Acsl4 | 2027 | 0.424 | 0.0785 | No |
| 7 | Msh2 | 3022 | 0.356 | 0.0594 | No |
| 8 | Ephx2 | 3451 | 0.331 | 0.0597 | No |
| 9 | Pex5 | 3452 | 0.331 | 0.0756 | No |
| 10 | Slc27a2 | 3880 | 0.313 | 0.0751 | No |
| 11 | Siah1a | 4131 | 0.299 | 0.0803 | No |
| 12 | Dhrs3 | 4245 | 0.293 | 0.0903 | No |
| 13 | Top2a | 4440 | 0.284 | 0.0968 | No |
| 14 | Dio1 | 4686 | 0.274 | 0.1011 | No |
| 15 | Abcb9 | 4932 | 0.263 | 0.1048 | No |
| 16 | Cacna1b | 5192 | 0.250 | 0.1073 | No |
| 17 | Itgb1bp1 | 5322 | 0.245 | 0.1144 | No |
| 18 | Isoc1 | 6054 | 0.219 | 0.0983 | No |
| 19 | Ercc1 | 6274 | 0.211 | 0.1004 | No |
| 20 | Smarcc1 | 7042 | 0.181 | 0.0813 | No |
| 21 | Crabp2 | 7372 | 0.168 | 0.0774 | No |
| 22 | Pex6 | 8020 | 0.149 | 0.0610 | No |
| 23 | Ide | 8500 | 0.137 | 0.0502 | No |
| 24 | Acsl5 | 8587 | 0.135 | 0.0535 | No |
| 25 | Mvp | 8699 | 0.131 | 0.0558 | No |
| 26 | Abcd2 | 8999 | 0.121 | 0.0507 | No |
| 27 | Abcc5 | 9023 | 0.120 | 0.0556 | No |
| 28 | Gnpat | 9273 | 0.113 | 0.0520 | No |
| 29 | Gstk1 | 9390 | 0.109 | 0.0530 | No |
| 30 | Cnbp | 9443 | 0.107 | 0.0563 | No |
| 31 | Eci2 | 9581 | 0.103 | 0.0562 | No |
| 32 | Nudt19 | 9588 | 0.102 | 0.0609 | No |
| 33 | Vps4b | 9657 | 0.100 | 0.0632 | No |
| 34 | Hmgcl | 9712 | 0.099 | 0.0660 | No |
| 35 | Abcd3 | 10028 | 0.089 | 0.0588 | No |
| 36 | Fads1 | 10033 | 0.089 | 0.0629 | No |
| 37 | Acot8 | 10294 | 0.081 | 0.0573 | No |
| 38 | Ercc3 | 10323 | 0.080 | 0.0602 | No |
| 39 | Ctbp1 | 10753 | 0.066 | 0.0478 | No |
| 40 | Ehhadh | 10991 | 0.059 | 0.0420 | No |
| 41 | Aldh9a1 | 11135 | 0.054 | 0.0394 | No |
| 42 | Crat | 11255 | 0.051 | 0.0375 | No |
| 43 | Elovl5 | 11573 | 0.041 | 0.0279 | No |
| 44 | Esr2 | 11580 | 0.041 | 0.0297 | No |
| 45 | Abcb4 | 11723 | 0.037 | 0.0263 | No |
| 46 | Dlg4 | 11794 | 0.034 | 0.0254 | No |
| 47 | Hsd17b4 | 12006 | 0.028 | 0.0191 | No |
| 48 | Abcb1a | 12183 | 0.023 | 0.0138 | No |
| 49 | Slc25a17 | 12352 | 0.018 | 0.0086 | No |
| 50 | Idi1 | 12409 | 0.017 | 0.0073 | No |
| 51 | Fabp6 | 12456 | 0.015 | 0.0064 | No |
| 52 | Hao2 | 12500 | 0.014 | 0.0055 | No |
| 53 | Hsd11b2 | 12600 | 0.012 | 0.0025 | No |
| 54 | Pex14 | 12686 | 0.009 | -0.0002 | No |
| 55 | Nr1i2 | 13006 | 0.000 | -0.0118 | No |
| 56 | Acaa1a | 14211 | -0.030 | -0.0541 | No |
| 57 | Pabpc1 | 14285 | -0.032 | -0.0552 | No |
| 58 | Acsl1 | 14454 | -0.037 | -0.0596 | No |
| 59 | Acox1 | 14997 | -0.053 | -0.0768 | No |
| 60 | Rdh11 | 15383 | -0.064 | -0.0877 | No |
| 61 | Sod1 | 15433 | -0.065 | -0.0863 | No |
| 62 | Retsat | 15993 | -0.083 | -0.1027 | No |
| 63 | Hras | 16044 | -0.085 | -0.1004 | No |
| 64 | Lonp2 | 16343 | -0.093 | -0.1068 | No |
| 65 | Cdk7 | 16611 | -0.100 | -0.1117 | No |
| 66 | Sod2 | 16692 | -0.102 | -0.1097 | No |
| 67 | Slc25a19 | 16950 | -0.111 | -0.1137 | No |
| 68 | Sema3c | 16984 | -0.113 | -0.1095 | No |
| 69 | Bcl10 | 17579 | -0.131 | -0.1248 | No |
| 70 | Pex2 | 17598 | -0.132 | -0.1191 | No |
| 71 | Scp2 | 18060 | -0.147 | -0.1288 | No |
| 72 | Abcc8 | 18242 | -0.151 | -0.1282 | No |
| 73 | Cadm1 | 18794 | -0.169 | -0.1401 | No |
| 74 | Fis1 | 18879 | -0.172 | -0.1349 | No |
| 75 | Idh1 | 19091 | -0.179 | -0.1340 | No |
| 76 | Atxn1 | 19378 | -0.188 | -0.1354 | No |
| 77 | Prdx1 | 19397 | -0.189 | -0.1270 | No |
| 78 | Ech1 | 19467 | -0.190 | -0.1204 | No |
| 79 | Slc23a2 | 19524 | -0.192 | -0.1132 | No |
| 80 | Pex13 | 19720 | -0.199 | -0.1107 | No |
| 81 | Slc35b2 | 19950 | -0.208 | -0.1091 | No |
| 82 | Pex11b | 19951 | -0.208 | -0.0991 | No |
| 83 | Idh2 | 20862 | -0.238 | -0.1208 | No |
| 84 | Slc25a4 | 22177 | -0.293 | -0.1545 | No |
| 85 | Rxrg | 22294 | -0.298 | -0.1444 | No |
| 86 | Abcd1 | 23983 | -0.377 | -0.1877 | Yes |
| 87 | Cat | 24330 | -0.396 | -0.1812 | Yes |
| 88 | Prdx5 | 24683 | -0.417 | -0.1740 | Yes |
| 89 | Mlycd | 24911 | -0.430 | -0.1617 | Yes |
| 90 | Cln6 | 25107 | -0.442 | -0.1475 | Yes |
| 91 | Fdps | 25219 | -0.450 | -0.1300 | Yes |
| 92 | Tspo | 26791 | -0.626 | -0.1571 | Yes |
| 93 | Pex11a | 26839 | -0.637 | -0.1282 | Yes |
| 94 | Dhcr24 | 26954 | -0.662 | -0.1006 | Yes |
| 95 | Hsd17b11 | 27241 | -0.750 | -0.0751 | Yes |
| 96 | Sult2b1 | 27361 | -0.806 | -0.0407 | Yes |
| 97 | Hsd3b7 | 27553 | -1.031 | 0.0018 | Yes |