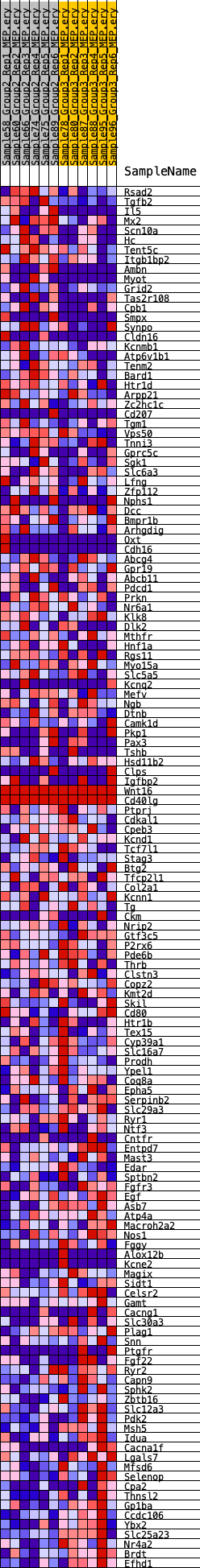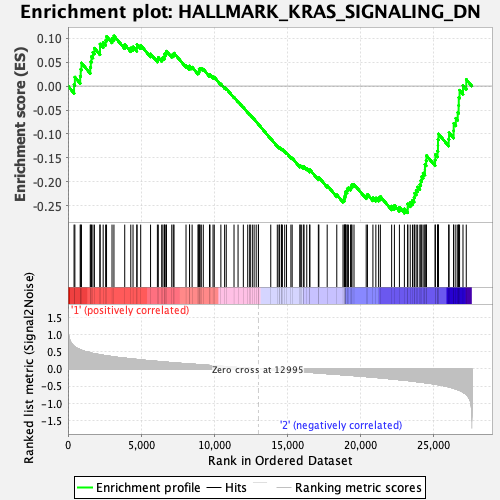
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.26522306 |
| Normalized Enrichment Score (NES) | -1.1294999 |
| Nominal p-value | 0.19277108 |
| FDR q-value | 0.78447026 |
| FWER p-Value | 0.977 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rsad2 | 414 | 0.656 | 0.0030 | No |
| 2 | Tgfb2 | 470 | 0.641 | 0.0187 | No |
| 3 | Il5 | 833 | 0.559 | 0.0210 | No |
| 4 | Mx2 | 870 | 0.552 | 0.0350 | No |
| 5 | Scn10a | 919 | 0.542 | 0.0482 | No |
| 6 | Hc | 1521 | 0.469 | 0.0393 | No |
| 7 | Tent5c | 1559 | 0.464 | 0.0507 | No |
| 8 | Itgb1bp2 | 1604 | 0.458 | 0.0618 | No |
| 9 | Ambn | 1703 | 0.447 | 0.0705 | No |
| 10 | Myot | 1805 | 0.441 | 0.0791 | No |
| 11 | Grid2 | 2187 | 0.409 | 0.0765 | No |
| 12 | Tas2r108 | 2188 | 0.409 | 0.0878 | No |
| 13 | Cpb1 | 2404 | 0.394 | 0.0908 | No |
| 14 | Smpx | 2573 | 0.384 | 0.0953 | No |
| 15 | Synpo | 2634 | 0.380 | 0.1036 | No |
| 16 | Cldn16 | 3009 | 0.356 | 0.0999 | No |
| 17 | Kcnmb1 | 3140 | 0.347 | 0.1047 | No |
| 18 | Atp6v1b1 | 3877 | 0.314 | 0.0866 | No |
| 19 | Tenm2 | 4292 | 0.291 | 0.0795 | No |
| 20 | Bard1 | 4445 | 0.284 | 0.0818 | No |
| 21 | Htr1d | 4706 | 0.273 | 0.0799 | No |
| 22 | Arpp21 | 4717 | 0.273 | 0.0871 | No |
| 23 | Zc2hc1c | 4969 | 0.261 | 0.0852 | No |
| 24 | Cd207 | 5643 | 0.231 | 0.0670 | No |
| 25 | Tgm1 | 6110 | 0.217 | 0.0561 | No |
| 26 | Vps50 | 6172 | 0.214 | 0.0598 | No |
| 27 | Tnni3 | 6405 | 0.207 | 0.0571 | No |
| 28 | Gprc5c | 6487 | 0.204 | 0.0597 | No |
| 29 | Sgk1 | 6591 | 0.200 | 0.0615 | No |
| 30 | Slc6a3 | 6600 | 0.200 | 0.0668 | No |
| 31 | Lfng | 6687 | 0.196 | 0.0690 | No |
| 32 | Zfp112 | 6731 | 0.194 | 0.0728 | No |
| 33 | Nphs1 | 7092 | 0.180 | 0.0647 | No |
| 34 | Dcc | 7194 | 0.176 | 0.0658 | No |
| 35 | Bmpr1b | 7247 | 0.173 | 0.0687 | No |
| 36 | Arhgdig | 8067 | 0.147 | 0.0430 | No |
| 37 | Oxt | 8306 | 0.140 | 0.0382 | No |
| 38 | Cdh16 | 8310 | 0.140 | 0.0419 | No |
| 39 | Abcg4 | 8484 | 0.138 | 0.0395 | No |
| 40 | Gpr19 | 8884 | 0.125 | 0.0284 | No |
| 41 | Abcb11 | 8940 | 0.123 | 0.0298 | No |
| 42 | Pdcd1 | 8962 | 0.122 | 0.0324 | No |
| 43 | Prkn | 8965 | 0.122 | 0.0357 | No |
| 44 | Nr6a1 | 9030 | 0.120 | 0.0367 | No |
| 45 | Klk8 | 9111 | 0.118 | 0.0370 | No |
| 46 | Dlk2 | 9248 | 0.114 | 0.0352 | No |
| 47 | Mthfr | 9672 | 0.100 | 0.0226 | No |
| 48 | Hnf1a | 9706 | 0.099 | 0.0241 | No |
| 49 | Rgs11 | 9914 | 0.092 | 0.0191 | No |
| 50 | Myo15a | 9998 | 0.090 | 0.0186 | No |
| 51 | Slc5a5 | 10449 | 0.077 | 0.0043 | No |
| 52 | Kcnq2 | 10706 | 0.068 | -0.0031 | No |
| 53 | Mefv | 10817 | 0.064 | -0.0053 | No |
| 54 | Ngb | 11348 | 0.048 | -0.0233 | No |
| 55 | Dtnb | 11624 | 0.039 | -0.0322 | No |
| 56 | Camk1d | 11985 | 0.029 | -0.0445 | No |
| 57 | Pkp1 | 12285 | 0.020 | -0.0549 | No |
| 58 | Pax3 | 12423 | 0.016 | -0.0594 | No |
| 59 | Tshb | 12457 | 0.015 | -0.0602 | No |
| 60 | Hsd11b2 | 12600 | 0.012 | -0.0650 | No |
| 61 | Clps | 12722 | 0.008 | -0.0692 | No |
| 62 | Igfbp2 | 12864 | 0.004 | -0.0742 | No |
| 63 | Wnt16 | 13016 | 0.000 | -0.0797 | No |
| 64 | Cd40lg | 13017 | 0.000 | -0.0797 | No |
| 65 | Ptprj | 13859 | -0.020 | -0.1098 | No |
| 66 | Cdkal1 | 14307 | -0.033 | -0.1252 | No |
| 67 | Cpeb3 | 14426 | -0.036 | -0.1285 | No |
| 68 | Kcnd1 | 14446 | -0.036 | -0.1282 | No |
| 69 | Tcf7l1 | 14586 | -0.041 | -0.1321 | No |
| 70 | Stag3 | 14588 | -0.041 | -0.1310 | No |
| 71 | Btg2 | 14654 | -0.043 | -0.1322 | No |
| 72 | Tfcp2l1 | 14812 | -0.048 | -0.1366 | No |
| 73 | Col2a1 | 14937 | -0.051 | -0.1397 | No |
| 74 | Kcnn1 | 15237 | -0.060 | -0.1489 | No |
| 75 | Tg | 15322 | -0.062 | -0.1502 | No |
| 76 | Ckm | 15846 | -0.078 | -0.1671 | No |
| 77 | Nrip2 | 15874 | -0.079 | -0.1660 | No |
| 78 | Gtf3c5 | 15973 | -0.082 | -0.1673 | No |
| 79 | P2rx6 | 16117 | -0.087 | -0.1701 | No |
| 80 | Pde6b | 16122 | -0.087 | -0.1678 | No |
| 81 | Thrb | 16320 | -0.093 | -0.1724 | No |
| 82 | Clstn3 | 16518 | -0.098 | -0.1769 | No |
| 83 | Copz2 | 16532 | -0.099 | -0.1746 | No |
| 84 | Kmt2d | 17112 | -0.116 | -0.1925 | No |
| 85 | Skil | 17148 | -0.117 | -0.1905 | No |
| 86 | Cd80 | 17716 | -0.136 | -0.2074 | No |
| 87 | Htr1b | 18372 | -0.156 | -0.2270 | No |
| 88 | Tex15 | 18798 | -0.169 | -0.2378 | No |
| 89 | Cyp39a1 | 18883 | -0.172 | -0.2361 | No |
| 90 | Slc16a7 | 18902 | -0.172 | -0.2320 | No |
| 91 | Prodh | 18939 | -0.174 | -0.2285 | No |
| 92 | Ypel1 | 18962 | -0.174 | -0.2245 | No |
| 93 | Coq8a | 18977 | -0.175 | -0.2202 | No |
| 94 | Epha5 | 19072 | -0.178 | -0.2187 | No |
| 95 | Serpinb2 | 19087 | -0.179 | -0.2142 | No |
| 96 | Slc29a3 | 19169 | -0.181 | -0.2122 | No |
| 97 | Ryr1 | 19314 | -0.186 | -0.2123 | No |
| 98 | Ntf3 | 19364 | -0.188 | -0.2089 | No |
| 99 | Cntfr | 19412 | -0.189 | -0.2054 | No |
| 100 | Entpd7 | 19549 | -0.193 | -0.2050 | No |
| 101 | Mast3 | 20383 | -0.222 | -0.2292 | No |
| 102 | Edar | 20469 | -0.225 | -0.2261 | No |
| 103 | Sptbn2 | 20842 | -0.237 | -0.2330 | No |
| 104 | Fgfr3 | 21043 | -0.246 | -0.2335 | No |
| 105 | Egf | 21232 | -0.254 | -0.2334 | No |
| 106 | Asb7 | 21352 | -0.259 | -0.2305 | No |
| 107 | Atp4a | 22122 | -0.291 | -0.2505 | No |
| 108 | Macroh2a2 | 22311 | -0.299 | -0.2491 | No |
| 109 | Nos1 | 22653 | -0.312 | -0.2529 | No |
| 110 | Fggy | 22993 | -0.326 | -0.2562 | Yes |
| 111 | Alox12b | 23222 | -0.337 | -0.2552 | Yes |
| 112 | Kcne2 | 23223 | -0.337 | -0.2459 | Yes |
| 113 | Magix | 23393 | -0.345 | -0.2425 | Yes |
| 114 | Sidt1 | 23546 | -0.354 | -0.2383 | Yes |
| 115 | Celsr2 | 23656 | -0.359 | -0.2324 | Yes |
| 116 | Gamt | 23693 | -0.361 | -0.2237 | Yes |
| 117 | Cacng1 | 23798 | -0.367 | -0.2173 | Yes |
| 118 | Slc30a3 | 23900 | -0.373 | -0.2107 | Yes |
| 119 | Plag1 | 24050 | -0.380 | -0.2056 | Yes |
| 120 | Snn | 24115 | -0.384 | -0.1974 | Yes |
| 121 | Ptgfr | 24176 | -0.387 | -0.1889 | Yes |
| 122 | Fgf22 | 24274 | -0.392 | -0.1816 | Yes |
| 123 | Ryr2 | 24398 | -0.401 | -0.1750 | Yes |
| 124 | Capn9 | 24400 | -0.401 | -0.1639 | Yes |
| 125 | Sphk2 | 24478 | -0.406 | -0.1555 | Yes |
| 126 | Zbtb16 | 24501 | -0.409 | -0.1450 | Yes |
| 127 | Slc12a3 | 25090 | -0.441 | -0.1542 | Yes |
| 128 | Pdk2 | 25111 | -0.442 | -0.1427 | Yes |
| 129 | Msh5 | 25259 | -0.454 | -0.1356 | Yes |
| 130 | Idua | 25288 | -0.456 | -0.1240 | Yes |
| 131 | Cacna1f | 25291 | -0.456 | -0.1114 | Yes |
| 132 | Lgals7 | 25322 | -0.459 | -0.0999 | Yes |
| 133 | Mfsd6 | 26021 | -0.520 | -0.1109 | Yes |
| 134 | Selenop | 26046 | -0.522 | -0.0974 | Yes |
| 135 | Cpa2 | 26358 | -0.561 | -0.0932 | Yes |
| 136 | Thnsl2 | 26365 | -0.561 | -0.0779 | Yes |
| 137 | Gp1ba | 26511 | -0.582 | -0.0671 | Yes |
| 138 | Ccdc106 | 26639 | -0.601 | -0.0551 | Yes |
| 139 | Ybx2 | 26701 | -0.610 | -0.0405 | Yes |
| 140 | Slc25a23 | 26711 | -0.612 | -0.0239 | Yes |
| 141 | Nr4a2 | 26765 | -0.622 | -0.0086 | Yes |
| 142 | Brdt | 26999 | -0.671 | 0.0014 | Yes |
| 143 | Efhd1 | 27223 | -0.742 | 0.0138 | Yes |