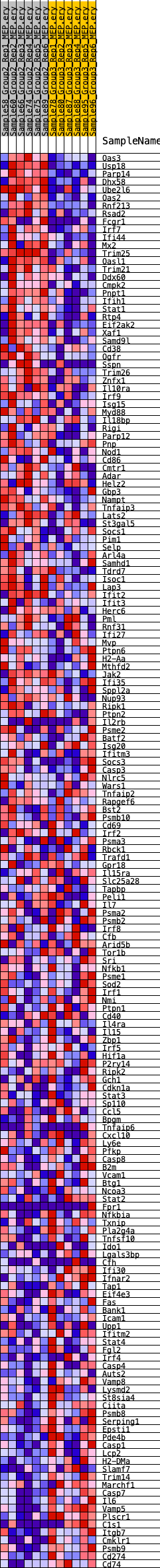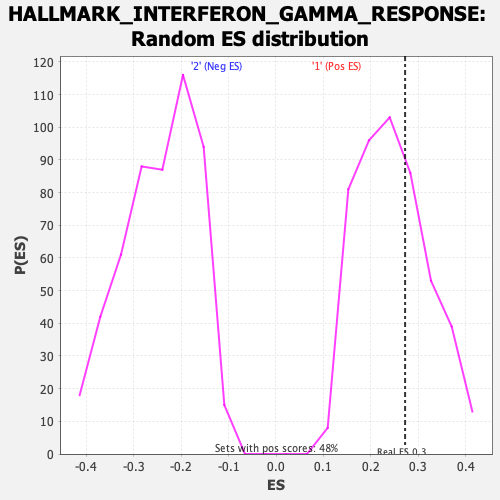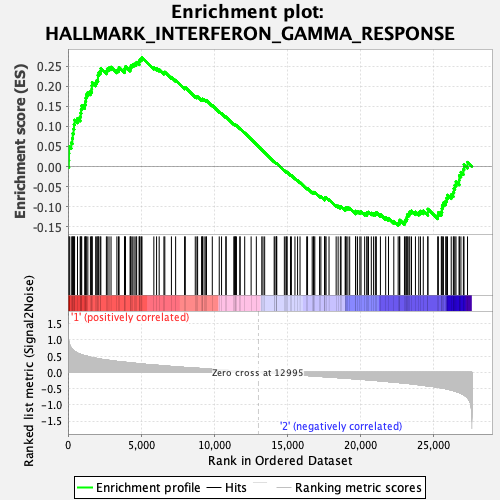
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_INTERFERON_GAMMA_RESPONSE |
| Enrichment Score (ES) | 0.272087 |
| Normalized Enrichment Score (NES) | 1.1051238 |
| Nominal p-value | 0.3361169 |
| FDR q-value | 0.5879584 |
| FWER p-Value | 0.979 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Oas3 | 60 | 0.932 | 0.0157 | Yes |
| 2 | Usp18 | 67 | 0.913 | 0.0331 | Yes |
| 3 | Parp14 | 74 | 0.902 | 0.0502 | Yes |
| 4 | Dhx58 | 221 | 0.748 | 0.0592 | Yes |
| 5 | Ube2l6 | 296 | 0.705 | 0.0701 | Yes |
| 6 | Oas2 | 324 | 0.694 | 0.0825 | Yes |
| 7 | Rnf213 | 380 | 0.669 | 0.0933 | Yes |
| 8 | Rsad2 | 414 | 0.656 | 0.1047 | Yes |
| 9 | Fcgr1 | 450 | 0.645 | 0.1159 | Yes |
| 10 | Irf7 | 649 | 0.590 | 0.1200 | Yes |
| 11 | Ifi44 | 831 | 0.560 | 0.1241 | Yes |
| 12 | Mx2 | 870 | 0.552 | 0.1334 | Yes |
| 13 | Trim25 | 898 | 0.547 | 0.1429 | Yes |
| 14 | Oasl1 | 937 | 0.540 | 0.1519 | Yes |
| 15 | Trim21 | 1142 | 0.510 | 0.1543 | Yes |
| 16 | Ddx60 | 1191 | 0.505 | 0.1622 | Yes |
| 17 | Cmpk2 | 1220 | 0.503 | 0.1709 | Yes |
| 18 | Pnpt1 | 1250 | 0.500 | 0.1794 | Yes |
| 19 | Ifih1 | 1356 | 0.492 | 0.1850 | Yes |
| 20 | Stat1 | 1533 | 0.468 | 0.1876 | Yes |
| 21 | Rtp4 | 1623 | 0.454 | 0.1931 | Yes |
| 22 | Eif2ak2 | 1629 | 0.454 | 0.2017 | Yes |
| 23 | Xaf1 | 1651 | 0.452 | 0.2096 | Yes |
| 24 | Samd9l | 1892 | 0.434 | 0.2092 | Yes |
| 25 | Cd38 | 1965 | 0.428 | 0.2148 | Yes |
| 26 | Ogfr | 2047 | 0.422 | 0.2199 | Yes |
| 27 | Sspn | 2048 | 0.422 | 0.2281 | Yes |
| 28 | Trim26 | 2104 | 0.418 | 0.2341 | Yes |
| 29 | Znfx1 | 2203 | 0.408 | 0.2384 | Yes |
| 30 | Il10ra | 2245 | 0.405 | 0.2446 | Yes |
| 31 | Irf9 | 2623 | 0.381 | 0.2382 | Yes |
| 32 | Isg15 | 2680 | 0.377 | 0.2434 | Yes |
| 33 | Myd88 | 2802 | 0.369 | 0.2461 | Yes |
| 34 | Il18bp | 2941 | 0.360 | 0.2480 | Yes |
| 35 | Rigi | 3342 | 0.337 | 0.2399 | Yes |
| 36 | Parp12 | 3449 | 0.331 | 0.2424 | Yes |
| 37 | Pnp | 3493 | 0.328 | 0.2471 | Yes |
| 38 | Nod1 | 3861 | 0.314 | 0.2398 | Yes |
| 39 | Cd86 | 3887 | 0.313 | 0.2449 | Yes |
| 40 | Cmtr1 | 3929 | 0.310 | 0.2494 | Yes |
| 41 | Adar | 4244 | 0.294 | 0.2436 | Yes |
| 42 | Helz2 | 4281 | 0.292 | 0.2479 | Yes |
| 43 | Gbp3 | 4315 | 0.289 | 0.2522 | Yes |
| 44 | Nampt | 4426 | 0.284 | 0.2537 | Yes |
| 45 | Tnfaip3 | 4518 | 0.279 | 0.2557 | Yes |
| 46 | Lats2 | 4640 | 0.276 | 0.2566 | Yes |
| 47 | St3gal5 | 4692 | 0.274 | 0.2600 | Yes |
| 48 | Socs1 | 4869 | 0.266 | 0.2587 | Yes |
| 49 | Pim1 | 4871 | 0.266 | 0.2638 | Yes |
| 50 | Selp | 4925 | 0.263 | 0.2669 | Yes |
| 51 | Arl4a | 5018 | 0.259 | 0.2685 | Yes |
| 52 | Samhd1 | 5057 | 0.257 | 0.2721 | Yes |
| 53 | Tdrd7 | 5868 | 0.224 | 0.2469 | No |
| 54 | Isoc1 | 6054 | 0.219 | 0.2443 | No |
| 55 | Lap3 | 6232 | 0.212 | 0.2419 | No |
| 56 | Ifit2 | 6557 | 0.201 | 0.2340 | No |
| 57 | Ifit3 | 6618 | 0.199 | 0.2356 | No |
| 58 | Herc6 | 7069 | 0.180 | 0.2227 | No |
| 59 | Pml | 7357 | 0.169 | 0.2155 | No |
| 60 | Rnf31 | 7968 | 0.151 | 0.1961 | No |
| 61 | Ifi27 | 8017 | 0.150 | 0.1973 | No |
| 62 | Mvp | 8699 | 0.131 | 0.1749 | No |
| 63 | Ptpn6 | 8813 | 0.127 | 0.1733 | No |
| 64 | H2-Aa | 8860 | 0.126 | 0.1740 | No |
| 65 | Mthfd2 | 9132 | 0.117 | 0.1664 | No |
| 66 | Jak2 | 9176 | 0.116 | 0.1671 | No |
| 67 | Ifi35 | 9198 | 0.115 | 0.1685 | No |
| 68 | Sppl2a | 9302 | 0.112 | 0.1669 | No |
| 69 | Nup93 | 9417 | 0.108 | 0.1648 | No |
| 70 | Ripk1 | 9465 | 0.106 | 0.1651 | No |
| 71 | Ptpn2 | 9863 | 0.094 | 0.1525 | No |
| 72 | Il2rb | 10338 | 0.080 | 0.1367 | No |
| 73 | Psme2 | 10496 | 0.075 | 0.1324 | No |
| 74 | Batf2 | 10790 | 0.065 | 0.1230 | No |
| 75 | Isg20 | 10810 | 0.064 | 0.1236 | No |
| 76 | Ifitm3 | 11334 | 0.048 | 0.1054 | No |
| 77 | Socs3 | 11387 | 0.047 | 0.1044 | No |
| 78 | Casp3 | 11434 | 0.045 | 0.1036 | No |
| 79 | Nlrc5 | 11443 | 0.045 | 0.1042 | No |
| 80 | Wars1 | 11514 | 0.043 | 0.1025 | No |
| 81 | Tnfaip2 | 11754 | 0.036 | 0.0944 | No |
| 82 | Rapgef6 | 11759 | 0.035 | 0.0950 | No |
| 83 | Bst2 | 12078 | 0.026 | 0.0839 | No |
| 84 | Psmb10 | 12517 | 0.014 | 0.0682 | No |
| 85 | Cd69 | 12873 | 0.004 | 0.0553 | No |
| 86 | Irf2 | 13247 | -0.003 | 0.0418 | No |
| 87 | Psma3 | 13329 | -0.005 | 0.0389 | No |
| 88 | Rbck1 | 13443 | -0.009 | 0.0350 | No |
| 89 | Trafd1 | 14093 | -0.027 | 0.0118 | No |
| 90 | Gpr18 | 14147 | -0.028 | 0.0104 | No |
| 91 | Il15ra | 14243 | -0.031 | 0.0075 | No |
| 92 | Slc25a28 | 14249 | -0.031 | 0.0080 | No |
| 93 | Tapbp | 14271 | -0.032 | 0.0078 | No |
| 94 | Peli1 | 14788 | -0.047 | -0.0101 | No |
| 95 | Il7 | 14894 | -0.050 | -0.0130 | No |
| 96 | Psma2 | 14946 | -0.052 | -0.0138 | No |
| 97 | Psmb2 | 14979 | -0.052 | -0.0140 | No |
| 98 | Irf8 | 15207 | -0.059 | -0.0211 | No |
| 99 | Cfb | 15270 | -0.061 | -0.0222 | No |
| 100 | Arid5b | 15513 | -0.068 | -0.0297 | No |
| 101 | Tor1b | 15692 | -0.073 | -0.0348 | No |
| 102 | Sri | 15859 | -0.078 | -0.0394 | No |
| 103 | Nfkb1 | 16316 | -0.092 | -0.0542 | No |
| 104 | Psme1 | 16374 | -0.094 | -0.0545 | No |
| 105 | Sod2 | 16692 | -0.102 | -0.0641 | No |
| 106 | Irf1 | 16782 | -0.106 | -0.0653 | No |
| 107 | Nmi | 16805 | -0.107 | -0.0641 | No |
| 108 | Ptpn1 | 16870 | -0.109 | -0.0643 | No |
| 109 | Cd40 | 17197 | -0.120 | -0.0739 | No |
| 110 | Il4ra | 17285 | -0.121 | -0.0747 | No |
| 111 | Il15 | 17541 | -0.130 | -0.0815 | No |
| 112 | Zbp1 | 17542 | -0.130 | -0.0790 | No |
| 113 | Irf5 | 17567 | -0.131 | -0.0774 | No |
| 114 | Hif1a | 17641 | -0.134 | -0.0775 | No |
| 115 | P2ry14 | 17842 | -0.139 | -0.0821 | No |
| 116 | Ripk2 | 18329 | -0.155 | -0.0968 | No |
| 117 | Gch1 | 18464 | -0.159 | -0.0987 | No |
| 118 | Cdkn1a | 18617 | -0.164 | -0.1011 | No |
| 119 | Stat3 | 18664 | -0.166 | -0.0995 | No |
| 120 | Sp110 | 18953 | -0.174 | -0.1067 | No |
| 121 | Ccl5 | 18958 | -0.174 | -0.1035 | No |
| 122 | Bpgm | 19021 | -0.176 | -0.1024 | No |
| 123 | Tnfaip6 | 19131 | -0.180 | -0.1029 | No |
| 124 | Cxcl10 | 19255 | -0.184 | -0.1038 | No |
| 125 | Ly6e | 19656 | -0.197 | -0.1146 | No |
| 126 | Pfkp | 19666 | -0.197 | -0.1111 | No |
| 127 | Casp8 | 19791 | -0.202 | -0.1118 | No |
| 128 | B2m | 19949 | -0.208 | -0.1135 | No |
| 129 | Vcam1 | 20029 | -0.211 | -0.1123 | No |
| 130 | Btg1 | 20278 | -0.218 | -0.1172 | No |
| 131 | Ncoa3 | 20426 | -0.224 | -0.1183 | No |
| 132 | Stat2 | 20443 | -0.224 | -0.1145 | No |
| 133 | Fpr1 | 20539 | -0.227 | -0.1136 | No |
| 134 | Nfkbia | 20732 | -0.232 | -0.1161 | No |
| 135 | Txnip | 20888 | -0.239 | -0.1172 | No |
| 136 | Pla2g4a | 21009 | -0.245 | -0.1169 | No |
| 137 | Tnfsf10 | 21075 | -0.247 | -0.1145 | No |
| 138 | Ido1 | 21354 | -0.259 | -0.1196 | No |
| 139 | Lgals3bp | 21714 | -0.273 | -0.1275 | No |
| 140 | Cfh | 21907 | -0.281 | -0.1291 | No |
| 141 | Ifi30 | 22269 | -0.297 | -0.1365 | No |
| 142 | Ifnar2 | 22568 | -0.308 | -0.1415 | No |
| 143 | Tap1 | 22656 | -0.312 | -0.1386 | No |
| 144 | Eif4e3 | 22681 | -0.313 | -0.1335 | No |
| 145 | Fas | 22998 | -0.327 | -0.1387 | No |
| 146 | Bank1 | 23045 | -0.329 | -0.1341 | No |
| 147 | Icam1 | 23118 | -0.332 | -0.1303 | No |
| 148 | Upp1 | 23171 | -0.335 | -0.1258 | No |
| 149 | Ifitm2 | 23189 | -0.336 | -0.1200 | No |
| 150 | Stat4 | 23300 | -0.339 | -0.1175 | No |
| 151 | Fgl2 | 23363 | -0.343 | -0.1131 | No |
| 152 | Irf4 | 23480 | -0.350 | -0.1106 | No |
| 153 | Casp4 | 23749 | -0.364 | -0.1134 | No |
| 154 | Auts2 | 23977 | -0.376 | -0.1144 | No |
| 155 | Vamp8 | 24091 | -0.382 | -0.1112 | No |
| 156 | Lysmd2 | 24284 | -0.393 | -0.1106 | No |
| 157 | St8sia4 | 24597 | -0.414 | -0.1141 | No |
| 158 | Ciita | 24598 | -0.414 | -0.1061 | No |
| 159 | Psmb8 | 25270 | -0.455 | -0.1218 | No |
| 160 | Serping1 | 25315 | -0.458 | -0.1146 | No |
| 161 | Epsti1 | 25517 | -0.473 | -0.1129 | No |
| 162 | Pde4b | 25560 | -0.476 | -0.1053 | No |
| 163 | Casp1 | 25583 | -0.478 | -0.0969 | No |
| 164 | Lcp2 | 25662 | -0.486 | -0.0904 | No |
| 165 | H2-DMa | 25817 | -0.500 | -0.0864 | No |
| 166 | Slamf7 | 25881 | -0.506 | -0.0789 | No |
| 167 | Trim14 | 25941 | -0.511 | -0.0713 | No |
| 168 | Marchf1 | 26194 | -0.538 | -0.0701 | No |
| 169 | Casp7 | 26339 | -0.558 | -0.0647 | No |
| 170 | Il6 | 26377 | -0.563 | -0.0552 | No |
| 171 | Vamp5 | 26443 | -0.572 | -0.0465 | No |
| 172 | Plscr1 | 26520 | -0.583 | -0.0381 | No |
| 173 | C1s1 | 26739 | -0.618 | -0.0342 | No |
| 174 | Itgb7 | 26746 | -0.619 | -0.0225 | No |
| 175 | Cmklr1 | 26857 | -0.641 | -0.0142 | No |
| 176 | Psmb9 | 27026 | -0.677 | -0.0073 | No |
| 177 | Cd274 | 27068 | -0.691 | 0.0045 | No |
| 178 | Cd74 | 27309 | -0.776 | 0.0107 | No |