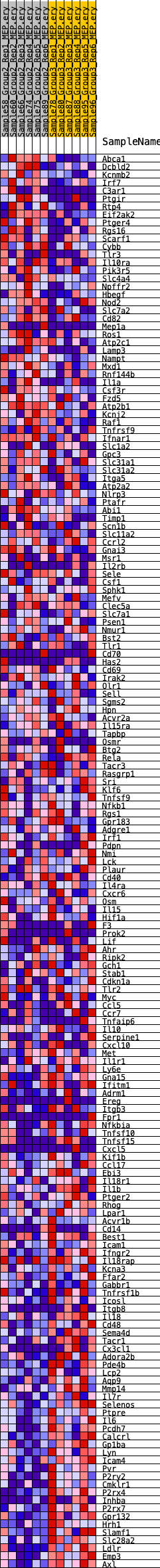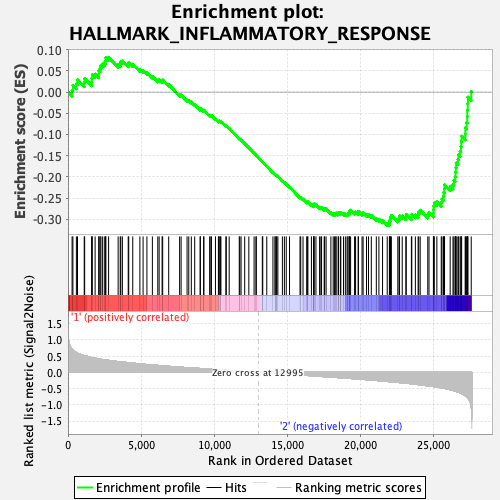
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
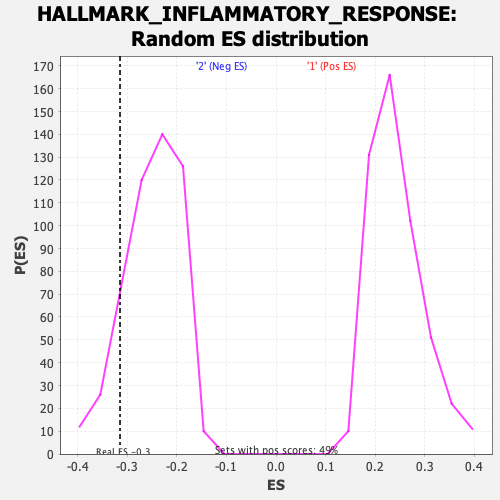
| Enrichment Score (ES) | -0.3149186 |
| Normalized Enrichment Score (NES) | -1.2695884 |
| Nominal p-value | 0.10453649 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.897 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 279 | 0.714 | 0.0043 | No |
| 2 | Dcbld2 | 335 | 0.687 | 0.0162 | No |
| 3 | Kcnmb2 | 574 | 0.609 | 0.0199 | No |
| 4 | Irf7 | 649 | 0.590 | 0.0291 | No |
| 5 | C3ar1 | 1109 | 0.513 | 0.0228 | No |
| 6 | Ptgir | 1139 | 0.510 | 0.0321 | No |
| 7 | Rtp4 | 1623 | 0.454 | 0.0237 | No |
| 8 | Eif2ak2 | 1629 | 0.454 | 0.0327 | No |
| 9 | Ptger4 | 1655 | 0.451 | 0.0409 | No |
| 10 | Rgs16 | 1853 | 0.438 | 0.0426 | No |
| 11 | Scarf1 | 2108 | 0.418 | 0.0418 | No |
| 12 | Cybb | 2109 | 0.418 | 0.0503 | No |
| 13 | Tlr3 | 2190 | 0.409 | 0.0556 | No |
| 14 | Il10ra | 2245 | 0.405 | 0.0619 | No |
| 15 | Pik3r5 | 2349 | 0.397 | 0.0662 | No |
| 16 | Slc4a4 | 2480 | 0.389 | 0.0693 | No |
| 17 | Npffr2 | 2570 | 0.384 | 0.0738 | No |
| 18 | Hbegf | 2585 | 0.383 | 0.0811 | No |
| 19 | Nod2 | 2774 | 0.371 | 0.0817 | No |
| 20 | Slc7a2 | 3426 | 0.331 | 0.0647 | No |
| 21 | Cd82 | 3569 | 0.326 | 0.0661 | No |
| 22 | Mep1a | 3607 | 0.324 | 0.0714 | No |
| 23 | Ros1 | 3716 | 0.321 | 0.0739 | No |
| 24 | Atp2c1 | 4116 | 0.300 | 0.0654 | No |
| 25 | Lamp3 | 4151 | 0.298 | 0.0702 | No |
| 26 | Nampt | 4426 | 0.284 | 0.0660 | No |
| 27 | Mxd1 | 4914 | 0.264 | 0.0536 | No |
| 28 | Rnf144b | 5132 | 0.254 | 0.0508 | No |
| 29 | Il1a | 5394 | 0.242 | 0.0462 | No |
| 30 | Csf3r | 5767 | 0.228 | 0.0373 | No |
| 31 | Fzd5 | 6138 | 0.215 | 0.0281 | No |
| 32 | Atp2b1 | 6229 | 0.212 | 0.0292 | No |
| 33 | Kcnj2 | 6431 | 0.206 | 0.0260 | No |
| 34 | Raf1 | 6473 | 0.205 | 0.0287 | No |
| 35 | Tnfrsf9 | 6883 | 0.188 | 0.0176 | No |
| 36 | Ifnar1 | 7629 | 0.163 | -0.0063 | No |
| 37 | Slc1a2 | 7732 | 0.159 | -0.0068 | No |
| 38 | Gpc3 | 8139 | 0.145 | -0.0187 | No |
| 39 | Slc31a1 | 8255 | 0.141 | -0.0200 | No |
| 40 | Slc31a2 | 8430 | 0.139 | -0.0235 | No |
| 41 | Itga5 | 8660 | 0.132 | -0.0292 | No |
| 42 | Atp2a2 | 9035 | 0.120 | -0.0404 | No |
| 43 | Nlrp3 | 9043 | 0.120 | -0.0382 | No |
| 44 | Ptafr | 9271 | 0.113 | -0.0442 | No |
| 45 | Abi1 | 9289 | 0.112 | -0.0426 | No |
| 46 | Timp1 | 9682 | 0.100 | -0.0548 | No |
| 47 | Scn1b | 9764 | 0.097 | -0.0558 | No |
| 48 | Slc11a2 | 9804 | 0.096 | -0.0553 | No |
| 49 | Ccrl2 | 10079 | 0.088 | -0.0635 | No |
| 50 | Gnai3 | 10281 | 0.081 | -0.0692 | No |
| 51 | Msr1 | 10309 | 0.080 | -0.0686 | No |
| 52 | Il2rb | 10338 | 0.080 | -0.0680 | No |
| 53 | Sele | 10417 | 0.077 | -0.0692 | No |
| 54 | Csf1 | 10453 | 0.077 | -0.0690 | No |
| 55 | Sphk1 | 10794 | 0.065 | -0.0800 | No |
| 56 | Mefv | 10817 | 0.064 | -0.0795 | No |
| 57 | Clec5a | 11017 | 0.059 | -0.0856 | No |
| 58 | Slc7a1 | 11706 | 0.037 | -0.1099 | No |
| 59 | Psen1 | 11742 | 0.036 | -0.1105 | No |
| 60 | Nmur1 | 11829 | 0.033 | -0.1129 | No |
| 61 | Bst2 | 12078 | 0.026 | -0.1215 | No |
| 62 | Tlr1 | 12360 | 0.018 | -0.1314 | No |
| 63 | Cd70 | 12720 | 0.008 | -0.1443 | No |
| 64 | Has2 | 12845 | 0.005 | -0.1487 | No |
| 65 | Cd69 | 12873 | 0.004 | -0.1496 | No |
| 66 | Irak2 | 13282 | -0.003 | -0.1644 | No |
| 67 | Olr1 | 13313 | -0.005 | -0.1654 | No |
| 68 | Sell | 13587 | -0.013 | -0.1751 | No |
| 69 | Sgms2 | 14009 | -0.024 | -0.1900 | No |
| 70 | Hpn | 14155 | -0.028 | -0.1947 | No |
| 71 | Acvr2a | 14157 | -0.028 | -0.1941 | No |
| 72 | Il15ra | 14243 | -0.031 | -0.1966 | No |
| 73 | Tapbp | 14271 | -0.032 | -0.1970 | No |
| 74 | Osmr | 14345 | -0.034 | -0.1989 | No |
| 75 | Btg2 | 14654 | -0.043 | -0.2093 | No |
| 76 | Rela | 14791 | -0.047 | -0.2133 | No |
| 77 | Tacr3 | 14918 | -0.051 | -0.2168 | No |
| 78 | Rasgrp1 | 15142 | -0.057 | -0.2238 | No |
| 79 | Sri | 15859 | -0.078 | -0.2483 | No |
| 80 | Klf6 | 15908 | -0.079 | -0.2485 | No |
| 81 | Tnfsf9 | 16065 | -0.085 | -0.2524 | No |
| 82 | Nfkb1 | 16316 | -0.092 | -0.2597 | No |
| 83 | Rgs1 | 16361 | -0.094 | -0.2594 | No |
| 84 | Gpr183 | 16397 | -0.095 | -0.2587 | No |
| 85 | Adgre1 | 16639 | -0.101 | -0.2655 | No |
| 86 | Irf1 | 16782 | -0.106 | -0.2685 | No |
| 87 | Pdpn | 16797 | -0.106 | -0.2669 | No |
| 88 | Nmi | 16805 | -0.107 | -0.2650 | No |
| 89 | Lck | 16824 | -0.107 | -0.2635 | No |
| 90 | Plaur | 16948 | -0.111 | -0.2657 | No |
| 91 | Cd40 | 17197 | -0.120 | -0.2723 | No |
| 92 | Il4ra | 17285 | -0.121 | -0.2730 | No |
| 93 | Cxcr6 | 17332 | -0.123 | -0.2722 | No |
| 94 | Osm | 17511 | -0.129 | -0.2761 | No |
| 95 | Il15 | 17541 | -0.130 | -0.2745 | No |
| 96 | Hif1a | 17641 | -0.134 | -0.2754 | No |
| 97 | F3 | 17969 | -0.143 | -0.2844 | No |
| 98 | Prok2 | 18127 | -0.149 | -0.2871 | No |
| 99 | Lif | 18211 | -0.150 | -0.2871 | No |
| 100 | Ahr | 18299 | -0.154 | -0.2872 | No |
| 101 | Ripk2 | 18329 | -0.155 | -0.2851 | No |
| 102 | Gch1 | 18464 | -0.159 | -0.2868 | No |
| 103 | Stab1 | 18490 | -0.160 | -0.2844 | No |
| 104 | Cdkn1a | 18617 | -0.164 | -0.2857 | No |
| 105 | Tlr2 | 18647 | -0.165 | -0.2834 | No |
| 106 | Myc | 18830 | -0.170 | -0.2866 | No |
| 107 | Ccl5 | 18958 | -0.174 | -0.2877 | No |
| 108 | Ccr7 | 19074 | -0.178 | -0.2883 | No |
| 109 | Tnfaip6 | 19131 | -0.180 | -0.2867 | No |
| 110 | Il10 | 19219 | -0.183 | -0.2861 | No |
| 111 | Serpine1 | 19224 | -0.183 | -0.2826 | No |
| 112 | Cxcl10 | 19255 | -0.184 | -0.2799 | No |
| 113 | Met | 19317 | -0.186 | -0.2784 | No |
| 114 | Il1r1 | 19588 | -0.194 | -0.2843 | No |
| 115 | Ly6e | 19656 | -0.197 | -0.2828 | No |
| 116 | Gna15 | 19840 | -0.204 | -0.2853 | No |
| 117 | Ifitm1 | 19844 | -0.205 | -0.2812 | No |
| 118 | Adrm1 | 20098 | -0.213 | -0.2862 | No |
| 119 | Ereg | 20172 | -0.216 | -0.2844 | No |
| 120 | Itgb3 | 20403 | -0.223 | -0.2883 | No |
| 121 | Fpr1 | 20539 | -0.227 | -0.2886 | No |
| 122 | Nfkbia | 20732 | -0.232 | -0.2909 | No |
| 123 | Tnfsf10 | 21075 | -0.247 | -0.2984 | No |
| 124 | Tnfsf15 | 21251 | -0.254 | -0.2996 | No |
| 125 | Cxcl5 | 21501 | -0.265 | -0.3033 | No |
| 126 | Kif1b | 21820 | -0.278 | -0.3093 | Yes |
| 127 | Ccl17 | 21968 | -0.284 | -0.3089 | Yes |
| 128 | Ebi3 | 21977 | -0.284 | -0.3034 | Yes |
| 129 | Il18r1 | 22023 | -0.287 | -0.2993 | Yes |
| 130 | Il1b | 22042 | -0.287 | -0.2941 | Yes |
| 131 | Ptger2 | 22104 | -0.290 | -0.2904 | Yes |
| 132 | Rhog | 22544 | -0.307 | -0.3002 | Yes |
| 133 | Lpar1 | 22630 | -0.310 | -0.2970 | Yes |
| 134 | Acvr1b | 22661 | -0.312 | -0.2918 | Yes |
| 135 | Cd14 | 22845 | -0.320 | -0.2920 | Yes |
| 136 | Best1 | 23096 | -0.331 | -0.2944 | Yes |
| 137 | Icam1 | 23118 | -0.332 | -0.2884 | Yes |
| 138 | Ifngr2 | 23471 | -0.349 | -0.2942 | Yes |
| 139 | Il18rap | 23500 | -0.351 | -0.2881 | Yes |
| 140 | Kcna3 | 23744 | -0.364 | -0.2896 | Yes |
| 141 | Ffar2 | 23932 | -0.374 | -0.2888 | Yes |
| 142 | Gabbr1 | 23975 | -0.376 | -0.2827 | Yes |
| 143 | Tnfrsf1b | 24074 | -0.381 | -0.2786 | Yes |
| 144 | Icosl | 24592 | -0.414 | -0.2891 | Yes |
| 145 | Itgb8 | 24677 | -0.417 | -0.2837 | Yes |
| 146 | Il18 | 24978 | -0.433 | -0.2858 | Yes |
| 147 | Cd48 | 24982 | -0.433 | -0.2772 | Yes |
| 148 | Sema4d | 24997 | -0.434 | -0.2689 | Yes |
| 149 | Tacr1 | 25036 | -0.437 | -0.2614 | Yes |
| 150 | Cx3cl1 | 25213 | -0.450 | -0.2587 | Yes |
| 151 | Adora2b | 25513 | -0.473 | -0.2600 | Yes |
| 152 | Pde4b | 25560 | -0.476 | -0.2521 | Yes |
| 153 | Lcp2 | 25662 | -0.486 | -0.2459 | Yes |
| 154 | Aqp9 | 25679 | -0.488 | -0.2366 | Yes |
| 155 | Mmp14 | 25726 | -0.492 | -0.2283 | Yes |
| 156 | Il7r | 25738 | -0.493 | -0.2187 | Yes |
| 157 | Selenos | 26123 | -0.530 | -0.2220 | Yes |
| 158 | Ptpre | 26315 | -0.555 | -0.2177 | Yes |
| 159 | Il6 | 26377 | -0.563 | -0.2085 | Yes |
| 160 | Pcdh7 | 26463 | -0.576 | -0.2000 | Yes |
| 161 | Calcrl | 26487 | -0.579 | -0.1891 | Yes |
| 162 | Gp1ba | 26511 | -0.582 | -0.1781 | Yes |
| 163 | Lyn | 26533 | -0.585 | -0.1670 | Yes |
| 164 | Icam4 | 26653 | -0.603 | -0.1592 | Yes |
| 165 | Pvr | 26684 | -0.607 | -0.1480 | Yes |
| 166 | P2ry2 | 26798 | -0.627 | -0.1394 | Yes |
| 167 | Cmklr1 | 26857 | -0.641 | -0.1285 | Yes |
| 168 | P2rx4 | 26878 | -0.645 | -0.1161 | Yes |
| 169 | Inhba | 26907 | -0.651 | -0.1040 | Yes |
| 170 | P2rx7 | 27155 | -0.717 | -0.0985 | Yes |
| 171 | Gpr132 | 27160 | -0.718 | -0.0841 | Yes |
| 172 | Hrh1 | 27246 | -0.754 | -0.0719 | Yes |
| 173 | Slamf1 | 27290 | -0.772 | -0.0578 | Yes |
| 174 | Slc28a2 | 27295 | -0.773 | -0.0423 | Yes |
| 175 | Ldlr | 27323 | -0.784 | -0.0274 | Yes |
| 176 | Emp3 | 27333 | -0.788 | -0.0117 | Yes |
| 177 | Axl | 27562 | -1.062 | 0.0015 | Yes |