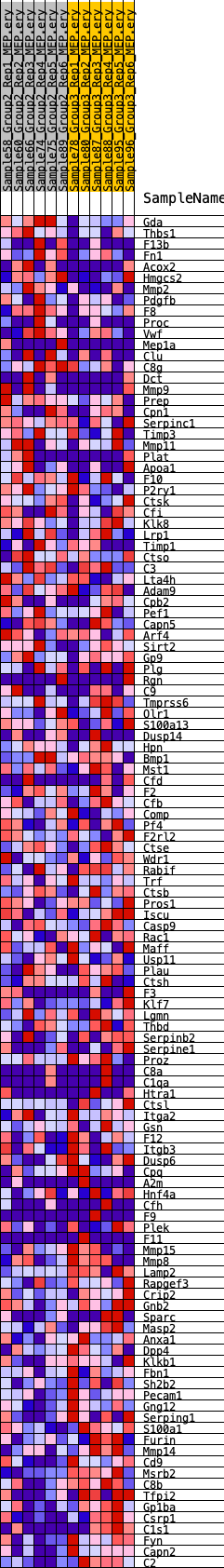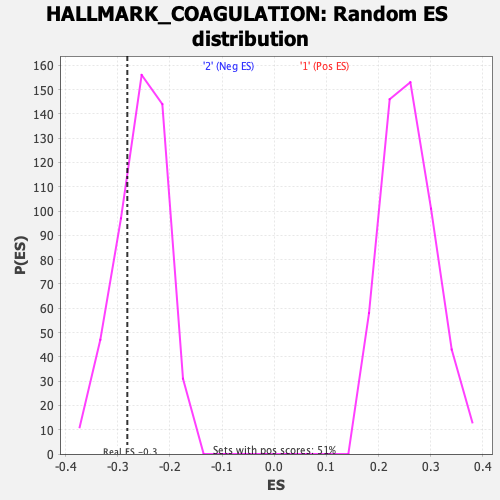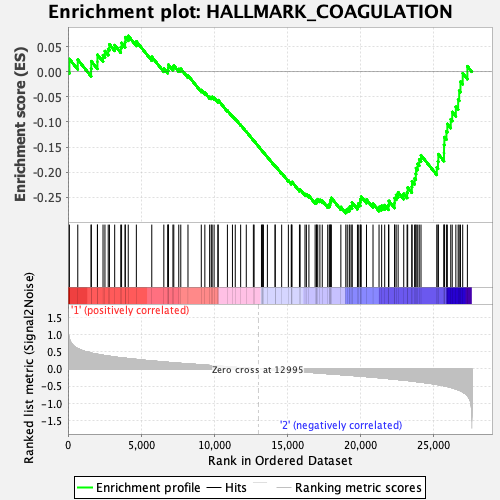
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.28161076 |
| Normalized Enrichment Score (NES) | -1.1026641 |
| Nominal p-value | 0.2674897 |
| FDR q-value | 0.6929639 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gda | 95 | 0.872 | 0.0254 | No |
| 2 | Thbs1 | 668 | 0.586 | 0.0241 | No |
| 3 | F13b | 1578 | 0.460 | 0.0062 | No |
| 4 | Fn1 | 1591 | 0.459 | 0.0210 | No |
| 5 | Acox2 | 2011 | 0.425 | 0.0199 | No |
| 6 | Hmgcs2 | 2013 | 0.425 | 0.0339 | No |
| 7 | Mmp2 | 2394 | 0.395 | 0.0332 | No |
| 8 | Pdgfb | 2525 | 0.386 | 0.0413 | No |
| 9 | F8 | 2749 | 0.372 | 0.0455 | No |
| 10 | Proc | 2832 | 0.367 | 0.0547 | No |
| 11 | Vwf | 3191 | 0.345 | 0.0531 | No |
| 12 | Mep1a | 3607 | 0.324 | 0.0487 | No |
| 13 | Clu | 3662 | 0.323 | 0.0575 | No |
| 14 | C8g | 3902 | 0.311 | 0.0591 | No |
| 15 | Dct | 3916 | 0.311 | 0.0689 | No |
| 16 | Mmp9 | 4118 | 0.299 | 0.0715 | No |
| 17 | Prep | 4672 | 0.275 | 0.0605 | No |
| 18 | Cpn1 | 5726 | 0.230 | 0.0298 | No |
| 19 | Serpinc1 | 6550 | 0.201 | 0.0065 | No |
| 20 | Timp3 | 6818 | 0.190 | 0.0031 | No |
| 21 | Mmp11 | 6853 | 0.189 | 0.0082 | No |
| 22 | Plat | 6860 | 0.189 | 0.0142 | No |
| 23 | Apoa1 | 7172 | 0.177 | 0.0088 | No |
| 24 | F10 | 7232 | 0.174 | 0.0124 | No |
| 25 | P2ry1 | 7571 | 0.165 | 0.0056 | No |
| 26 | Ctsk | 7709 | 0.160 | 0.0059 | No |
| 27 | Cfi | 8201 | 0.143 | -0.0072 | No |
| 28 | Klk8 | 9111 | 0.118 | -0.0364 | No |
| 29 | Lrp1 | 9354 | 0.110 | -0.0415 | No |
| 30 | Timp1 | 9682 | 0.100 | -0.0501 | No |
| 31 | Ctso | 9792 | 0.096 | -0.0509 | No |
| 32 | C3 | 9852 | 0.094 | -0.0499 | No |
| 33 | Lta4h | 9987 | 0.090 | -0.0518 | No |
| 34 | Adam9 | 10244 | 0.082 | -0.0584 | No |
| 35 | Cpb2 | 10267 | 0.081 | -0.0565 | No |
| 36 | Pef1 | 10896 | 0.062 | -0.0773 | No |
| 37 | Capn5 | 11241 | 0.051 | -0.0881 | No |
| 38 | Arf4 | 11433 | 0.045 | -0.0936 | No |
| 39 | Sirt2 | 11801 | 0.034 | -0.1058 | No |
| 40 | Gp9 | 12186 | 0.023 | -0.1190 | No |
| 41 | Plg | 12671 | 0.010 | -0.1363 | No |
| 42 | Rgn | 12721 | 0.008 | -0.1378 | No |
| 43 | C9 | 13224 | -0.002 | -0.1560 | No |
| 44 | Tmprss6 | 13279 | -0.003 | -0.1579 | No |
| 45 | Olr1 | 13313 | -0.005 | -0.1589 | No |
| 46 | S100a13 | 13356 | -0.006 | -0.1602 | No |
| 47 | Dusp14 | 13629 | -0.014 | -0.1697 | No |
| 48 | Hpn | 14155 | -0.028 | -0.1878 | No |
| 49 | Bmp1 | 14162 | -0.028 | -0.1871 | No |
| 50 | Mst1 | 14605 | -0.041 | -0.2018 | No |
| 51 | Cfd | 15063 | -0.055 | -0.2166 | No |
| 52 | F2 | 15266 | -0.061 | -0.2220 | No |
| 53 | Cfb | 15270 | -0.061 | -0.2201 | No |
| 54 | Comp | 15312 | -0.062 | -0.2195 | No |
| 55 | Pf4 | 15821 | -0.077 | -0.2354 | No |
| 56 | F2rl2 | 15865 | -0.078 | -0.2344 | No |
| 57 | Ctse | 16200 | -0.089 | -0.2436 | No |
| 58 | Wdr1 | 16295 | -0.092 | -0.2440 | No |
| 59 | Rabif | 16467 | -0.097 | -0.2470 | No |
| 60 | Trf | 16893 | -0.109 | -0.2589 | No |
| 61 | Ctsb | 16985 | -0.113 | -0.2584 | No |
| 62 | Pros1 | 17005 | -0.113 | -0.2554 | No |
| 63 | Iscu | 17071 | -0.115 | -0.2539 | No |
| 64 | Casp9 | 17218 | -0.120 | -0.2552 | No |
| 65 | Rac1 | 17380 | -0.125 | -0.2570 | No |
| 66 | Maff | 17752 | -0.137 | -0.2659 | No |
| 67 | Usp11 | 17857 | -0.140 | -0.2650 | No |
| 68 | Plau | 17905 | -0.141 | -0.2621 | No |
| 69 | Ctsh | 17928 | -0.142 | -0.2582 | No |
| 70 | F3 | 17969 | -0.143 | -0.2549 | No |
| 71 | Klf7 | 17997 | -0.144 | -0.2511 | No |
| 72 | Lgmn | 18651 | -0.165 | -0.2693 | No |
| 73 | Thbd | 18989 | -0.175 | -0.2758 | Yes |
| 74 | Serpinb2 | 19087 | -0.179 | -0.2734 | Yes |
| 75 | Serpine1 | 19224 | -0.183 | -0.2723 | Yes |
| 76 | Proz | 19275 | -0.185 | -0.2680 | Yes |
| 77 | C8a | 19413 | -0.189 | -0.2667 | Yes |
| 78 | C1qa | 19414 | -0.189 | -0.2604 | Yes |
| 79 | Htra1 | 19785 | -0.202 | -0.2672 | Yes |
| 80 | Ctsl | 19843 | -0.204 | -0.2625 | Yes |
| 81 | Itga2 | 19974 | -0.209 | -0.2603 | Yes |
| 82 | Gsn | 19982 | -0.209 | -0.2536 | Yes |
| 83 | F12 | 20040 | -0.211 | -0.2487 | Yes |
| 84 | Itgb3 | 20403 | -0.223 | -0.2545 | Yes |
| 85 | Dusp6 | 20849 | -0.238 | -0.2628 | Yes |
| 86 | Cpq | 21261 | -0.255 | -0.2693 | Yes |
| 87 | A2m | 21439 | -0.263 | -0.2670 | Yes |
| 88 | Hnf4a | 21648 | -0.270 | -0.2656 | Yes |
| 89 | Cfh | 21907 | -0.281 | -0.2657 | Yes |
| 90 | F9 | 21933 | -0.282 | -0.2572 | Yes |
| 91 | Plek | 22324 | -0.300 | -0.2615 | Yes |
| 92 | F11 | 22329 | -0.300 | -0.2517 | Yes |
| 93 | Mmp15 | 22430 | -0.302 | -0.2453 | Yes |
| 94 | Mmp8 | 22566 | -0.308 | -0.2400 | Yes |
| 95 | Lamp2 | 22948 | -0.324 | -0.2431 | Yes |
| 96 | Rapgef3 | 23179 | -0.335 | -0.2404 | Yes |
| 97 | Crip2 | 23218 | -0.336 | -0.2306 | Yes |
| 98 | Gnb2 | 23503 | -0.351 | -0.2293 | Yes |
| 99 | Sparc | 23515 | -0.352 | -0.2180 | Yes |
| 100 | Masp2 | 23692 | -0.361 | -0.2125 | Yes |
| 101 | Anxa1 | 23774 | -0.365 | -0.2033 | Yes |
| 102 | Dpp4 | 23791 | -0.366 | -0.1917 | Yes |
| 103 | Klkb1 | 23905 | -0.373 | -0.1835 | Yes |
| 104 | Fbn1 | 24009 | -0.379 | -0.1747 | Yes |
| 105 | Sh2b2 | 24129 | -0.384 | -0.1662 | Yes |
| 106 | Pecam1 | 25207 | -0.449 | -0.1905 | Yes |
| 107 | Gng12 | 25289 | -0.456 | -0.1784 | Yes |
| 108 | Serping1 | 25315 | -0.458 | -0.1641 | Yes |
| 109 | S100a1 | 25700 | -0.490 | -0.1618 | Yes |
| 110 | Furin | 25704 | -0.490 | -0.1457 | Yes |
| 111 | Mmp14 | 25726 | -0.492 | -0.1302 | Yes |
| 112 | Cd9 | 25866 | -0.504 | -0.1185 | Yes |
| 113 | Msrb2 | 25929 | -0.510 | -0.1039 | Yes |
| 114 | C8b | 26163 | -0.534 | -0.0946 | Yes |
| 115 | Tfpi2 | 26262 | -0.547 | -0.0801 | Yes |
| 116 | Gp1ba | 26511 | -0.582 | -0.0698 | Yes |
| 117 | Csrp1 | 26668 | -0.604 | -0.0554 | Yes |
| 118 | C1s1 | 26739 | -0.618 | -0.0375 | Yes |
| 119 | Fyn | 26827 | -0.635 | -0.0196 | Yes |
| 120 | Capn2 | 26980 | -0.666 | -0.0031 | Yes |
| 121 | C2 | 27299 | -0.774 | 0.0110 | Yes |