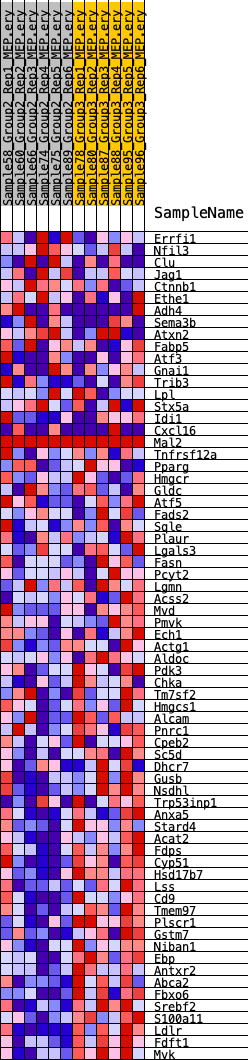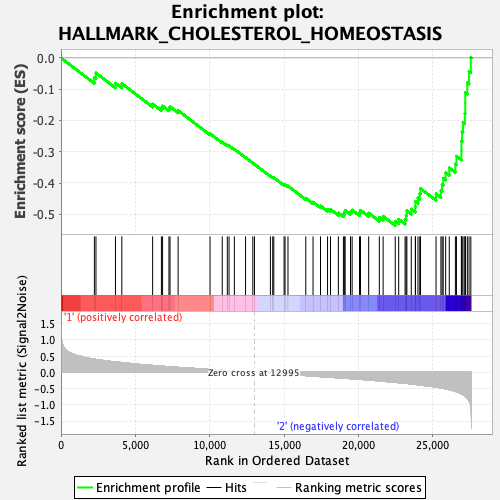
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.5361814 |
| Normalized Enrichment Score (NES) | -1.5937866 |
| Nominal p-value | 0.03206413 |
| FDR q-value | 0.9722361 |
| FWER p-Value | 0.376 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Errfi1 | 2244 | 0.405 | -0.0627 | No |
| 2 | Nfil3 | 2351 | 0.397 | -0.0480 | No |
| 3 | Clu | 3662 | 0.323 | -0.0806 | No |
| 4 | Jag1 | 4090 | 0.301 | -0.0821 | No |
| 5 | Ctnnb1 | 6162 | 0.215 | -0.1473 | No |
| 6 | Ethe1 | 6751 | 0.193 | -0.1597 | No |
| 7 | Adh4 | 6830 | 0.190 | -0.1537 | No |
| 8 | Sema3b | 7259 | 0.173 | -0.1612 | No |
| 9 | Atxn2 | 7331 | 0.170 | -0.1558 | No |
| 10 | Fabp5 | 7875 | 0.154 | -0.1684 | No |
| 11 | Atf3 | 10021 | 0.089 | -0.2422 | No |
| 12 | Gnai1 | 10843 | 0.063 | -0.2690 | No |
| 13 | Trib3 | 11183 | 0.053 | -0.2789 | No |
| 14 | Lpl | 11300 | 0.049 | -0.2808 | No |
| 15 | Stx5a | 11657 | 0.039 | -0.2919 | No |
| 16 | Idi1 | 12409 | 0.017 | -0.3184 | No |
| 17 | Cxcl16 | 12899 | 0.003 | -0.3360 | No |
| 18 | Mal2 | 13009 | 0.000 | -0.3400 | No |
| 19 | Tnfrsf12a | 14081 | -0.026 | -0.3776 | No |
| 20 | Pparg | 14237 | -0.031 | -0.3818 | No |
| 21 | Hmgcr | 14309 | -0.033 | -0.3829 | No |
| 22 | Gldc | 15000 | -0.053 | -0.4055 | No |
| 23 | Atf5 | 15065 | -0.055 | -0.4052 | No |
| 24 | Fads2 | 15263 | -0.061 | -0.4096 | No |
| 25 | Sqle | 16459 | -0.097 | -0.4485 | No |
| 26 | Plaur | 16948 | -0.111 | -0.4610 | No |
| 27 | Lgals3 | 17450 | -0.127 | -0.4733 | No |
| 28 | Fasn | 17924 | -0.142 | -0.4839 | No |
| 29 | Pcyt2 | 18122 | -0.149 | -0.4841 | No |
| 30 | Lgmn | 18651 | -0.165 | -0.4956 | No |
| 31 | Acss2 | 18990 | -0.175 | -0.4997 | No |
| 32 | Mvd | 19052 | -0.178 | -0.4937 | No |
| 33 | Pmvk | 19107 | -0.180 | -0.4873 | No |
| 34 | Ech1 | 19467 | -0.190 | -0.4915 | No |
| 35 | Actg1 | 19577 | -0.194 | -0.4864 | No |
| 36 | Aldoc | 20072 | -0.212 | -0.4945 | No |
| 37 | Pdk3 | 20141 | -0.215 | -0.4870 | No |
| 38 | Chka | 20689 | -0.230 | -0.4961 | No |
| 39 | Tm7sf2 | 21405 | -0.261 | -0.5099 | No |
| 40 | Hmgcs1 | 21660 | -0.270 | -0.5066 | No |
| 41 | Alcam | 22476 | -0.305 | -0.5220 | Yes |
| 42 | Pnrc1 | 22707 | -0.314 | -0.5157 | Yes |
| 43 | Cpeb2 | 23136 | -0.333 | -0.5158 | Yes |
| 44 | Sc5d | 23204 | -0.336 | -0.5026 | Yes |
| 45 | Dhcr7 | 23257 | -0.337 | -0.4888 | Yes |
| 46 | Gusb | 23554 | -0.354 | -0.4831 | Yes |
| 47 | Nsdhl | 23828 | -0.369 | -0.4759 | Yes |
| 48 | Trp53inp1 | 23834 | -0.369 | -0.4589 | Yes |
| 49 | Anxa5 | 24004 | -0.378 | -0.4474 | Yes |
| 50 | Stard4 | 24125 | -0.384 | -0.4339 | Yes |
| 51 | Acat2 | 24174 | -0.387 | -0.4177 | Yes |
| 52 | Fdps | 25219 | -0.450 | -0.4346 | Yes |
| 53 | Cyp51 | 25548 | -0.475 | -0.4245 | Yes |
| 54 | Hsd17b7 | 25626 | -0.483 | -0.4048 | Yes |
| 55 | Lss | 25701 | -0.490 | -0.3847 | Yes |
| 56 | Cd9 | 25866 | -0.504 | -0.3672 | Yes |
| 57 | Tmem97 | 26109 | -0.529 | -0.3514 | Yes |
| 58 | Plscr1 | 26520 | -0.583 | -0.3392 | Yes |
| 59 | Gstm7 | 26591 | -0.594 | -0.3141 | Yes |
| 60 | Niban1 | 26926 | -0.655 | -0.2958 | Yes |
| 61 | Ebp | 26927 | -0.655 | -0.2653 | Yes |
| 62 | Antxr2 | 26968 | -0.664 | -0.2359 | Yes |
| 63 | Abca2 | 27033 | -0.680 | -0.2065 | Yes |
| 64 | Fbxo6 | 27157 | -0.717 | -0.1776 | Yes |
| 65 | Srebf2 | 27184 | -0.728 | -0.1447 | Yes |
| 66 | S100a11 | 27185 | -0.729 | -0.1108 | Yes |
| 67 | Ldlr | 27323 | -0.784 | -0.0793 | Yes |
| 68 | Fdft1 | 27439 | -0.866 | -0.0432 | Yes |
| 69 | Mvk | 27559 | -1.055 | 0.0016 | Yes |