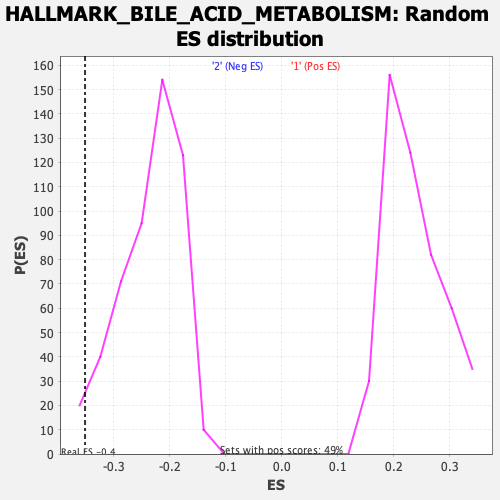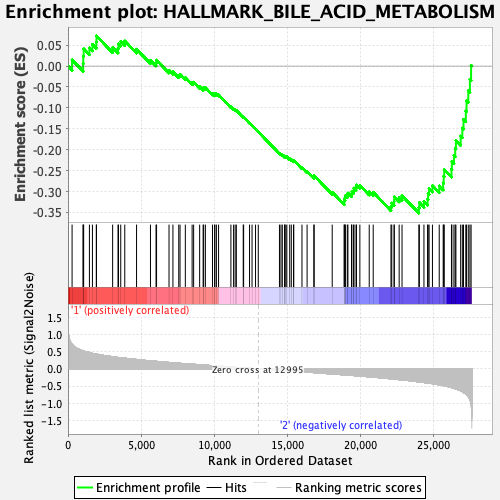
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.35165688 |
| Normalized Enrichment Score (NES) | -1.4928162 |
| Nominal p-value | 0.01754386 |
| FDR q-value | 0.8812053 |
| FWER p-Value | 0.547 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 279 | 0.714 | 0.0149 | No |
| 2 | Pfkm | 1032 | 0.524 | 0.0060 | No |
| 3 | Abca6 | 1043 | 0.523 | 0.0240 | No |
| 4 | Aldh1a1 | 1070 | 0.518 | 0.0412 | No |
| 5 | Slc29a1 | 1464 | 0.478 | 0.0437 | No |
| 6 | Abcg8 | 1670 | 0.450 | 0.0520 | No |
| 7 | Abca5 | 1936 | 0.430 | 0.0575 | No |
| 8 | Sult1b1 | 1945 | 0.430 | 0.0723 | No |
| 9 | Slc27a5 | 3051 | 0.354 | 0.0445 | No |
| 10 | Slco1a4 | 3419 | 0.332 | 0.0428 | No |
| 11 | Ephx2 | 3451 | 0.331 | 0.0533 | No |
| 12 | Amacr | 3605 | 0.324 | 0.0591 | No |
| 13 | Slc27a2 | 3880 | 0.313 | 0.0601 | No |
| 14 | Dio1 | 4686 | 0.274 | 0.0404 | No |
| 15 | Pipox | 5637 | 0.231 | 0.0140 | No |
| 16 | Nr0b2 | 6018 | 0.220 | 0.0079 | No |
| 17 | Isoc1 | 6054 | 0.219 | 0.0143 | No |
| 18 | Akr1d1 | 6905 | 0.188 | -0.0100 | No |
| 19 | Apoa1 | 7172 | 0.177 | -0.0135 | No |
| 20 | Aldh8a1 | 7570 | 0.165 | -0.0221 | No |
| 21 | Gclm | 7658 | 0.162 | -0.0196 | No |
| 22 | Pex6 | 8020 | 0.149 | -0.0275 | No |
| 23 | Abcg4 | 8484 | 0.138 | -0.0395 | No |
| 24 | Acsl5 | 8587 | 0.135 | -0.0385 | No |
| 25 | Abcd2 | 8999 | 0.121 | -0.0492 | No |
| 26 | Abca3 | 9236 | 0.115 | -0.0537 | No |
| 27 | Gnpat | 9273 | 0.113 | -0.0511 | No |
| 28 | Gstk1 | 9390 | 0.109 | -0.0515 | No |
| 29 | Nr1h4 | 9876 | 0.093 | -0.0658 | No |
| 30 | Abcd3 | 10028 | 0.089 | -0.0682 | No |
| 31 | Fads1 | 10033 | 0.089 | -0.0652 | No |
| 32 | Nedd4 | 10144 | 0.085 | -0.0662 | No |
| 33 | Dio2 | 10292 | 0.081 | -0.0688 | No |
| 34 | Aldh9a1 | 11135 | 0.054 | -0.0975 | No |
| 35 | Abca8b | 11319 | 0.049 | -0.1024 | No |
| 36 | Pxmp2 | 11424 | 0.046 | -0.1046 | No |
| 37 | Bcar3 | 11507 | 0.043 | -0.1061 | No |
| 38 | Bbox1 | 11977 | 0.029 | -0.1221 | No |
| 39 | Hsd17b4 | 12006 | 0.028 | -0.1221 | No |
| 40 | Idi1 | 12409 | 0.017 | -0.1362 | No |
| 41 | Slc23a1 | 12580 | 0.012 | -0.1419 | No |
| 42 | Pex1 | 12819 | 0.006 | -0.1504 | No |
| 43 | Nr1i2 | 13006 | 0.000 | -0.1571 | No |
| 44 | Acsl1 | 14454 | -0.037 | -0.2085 | No |
| 45 | Fdxr | 14557 | -0.040 | -0.2108 | No |
| 46 | Paox | 14649 | -0.043 | -0.2126 | No |
| 47 | Abca4 | 14806 | -0.048 | -0.2166 | No |
| 48 | Tfcp2l1 | 14812 | -0.048 | -0.2151 | No |
| 49 | Pnpla8 | 14890 | -0.050 | -0.2161 | No |
| 50 | Nudt12 | 14959 | -0.052 | -0.2168 | No |
| 51 | Pex19 | 15155 | -0.057 | -0.2218 | No |
| 52 | Fads2 | 15263 | -0.061 | -0.2236 | No |
| 53 | Bmp6 | 15413 | -0.065 | -0.2268 | No |
| 54 | Sod1 | 15433 | -0.065 | -0.2252 | No |
| 55 | Retsat | 15993 | -0.083 | -0.2426 | No |
| 56 | Lonp2 | 16343 | -0.093 | -0.2520 | No |
| 57 | Crot | 16807 | -0.107 | -0.2651 | No |
| 58 | Lck | 16824 | -0.107 | -0.2619 | No |
| 59 | Scp2 | 18060 | -0.147 | -0.3017 | No |
| 60 | Cyp39a1 | 18883 | -0.172 | -0.3255 | No |
| 61 | Pex7 | 18884 | -0.172 | -0.3195 | No |
| 62 | Optn | 18935 | -0.173 | -0.3152 | No |
| 63 | Npc1 | 18959 | -0.174 | -0.3100 | No |
| 64 | Idh1 | 19091 | -0.179 | -0.3084 | No |
| 65 | Hao1 | 19134 | -0.180 | -0.3036 | No |
| 66 | Atxn1 | 19378 | -0.188 | -0.3059 | No |
| 67 | Klf1 | 19403 | -0.189 | -0.3001 | No |
| 68 | Slc23a2 | 19524 | -0.192 | -0.2977 | No |
| 69 | Cyp27a1 | 19545 | -0.193 | -0.2917 | No |
| 70 | Rbp1 | 19687 | -0.198 | -0.2899 | No |
| 71 | Pex13 | 19720 | -0.199 | -0.2841 | No |
| 72 | Slc35b2 | 19950 | -0.208 | -0.2851 | No |
| 73 | Pex26 | 20590 | -0.228 | -0.3003 | No |
| 74 | Idh2 | 20862 | -0.238 | -0.3018 | No |
| 75 | Abca9 | 22077 | -0.289 | -0.3359 | No |
| 76 | Rxra | 22130 | -0.291 | -0.3275 | No |
| 77 | Rxrg | 22294 | -0.298 | -0.3230 | No |
| 78 | Pex16 | 22298 | -0.299 | -0.3126 | No |
| 79 | Gnmt | 22636 | -0.311 | -0.3140 | No |
| 80 | Phyh | 22832 | -0.319 | -0.3098 | No |
| 81 | Abcd1 | 23983 | -0.377 | -0.3384 | Yes |
| 82 | Lipe | 24016 | -0.379 | -0.3263 | Yes |
| 83 | Cat | 24330 | -0.396 | -0.3238 | Yes |
| 84 | Nr3c2 | 24578 | -0.413 | -0.3183 | Yes |
| 85 | Soat2 | 24616 | -0.415 | -0.3051 | Yes |
| 86 | Prdx5 | 24683 | -0.417 | -0.2928 | Yes |
| 87 | Mlycd | 24911 | -0.430 | -0.2860 | Yes |
| 88 | Hacl1 | 25375 | -0.463 | -0.2866 | Yes |
| 89 | Cyp7b1 | 25645 | -0.484 | -0.2794 | Yes |
| 90 | Aqp9 | 25679 | -0.488 | -0.2634 | Yes |
| 91 | Pecr | 25716 | -0.491 | -0.2475 | Yes |
| 92 | Pex11g | 26211 | -0.540 | -0.2465 | Yes |
| 93 | Pex12 | 26239 | -0.543 | -0.2284 | Yes |
| 94 | Gc | 26380 | -0.563 | -0.2138 | Yes |
| 95 | Ar | 26469 | -0.576 | -0.1967 | Yes |
| 96 | Cyp46a1 | 26515 | -0.583 | -0.1779 | Yes |
| 97 | Pex11a | 26839 | -0.637 | -0.1673 | Yes |
| 98 | Dhcr24 | 26954 | -0.662 | -0.1482 | Yes |
| 99 | Abca2 | 27033 | -0.680 | -0.1272 | Yes |
| 100 | Efhc1 | 27193 | -0.730 | -0.1074 | Yes |
| 101 | Hsd17b11 | 27241 | -0.750 | -0.0827 | Yes |
| 102 | Sult2b1 | 27361 | -0.806 | -0.0588 | Yes |
| 103 | Slc22a18 | 27472 | -0.892 | -0.0315 | Yes |
| 104 | Hsd3b7 | 27553 | -1.031 | 0.0018 | Yes |