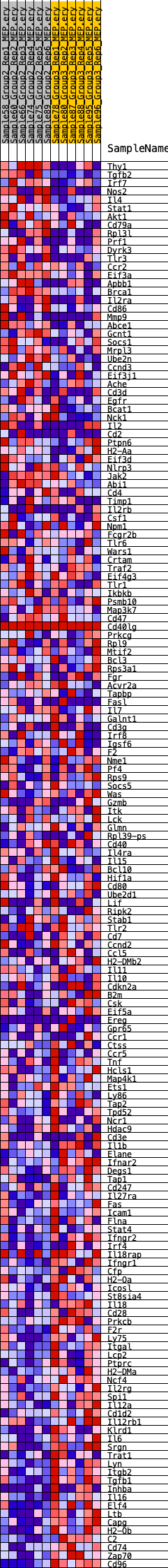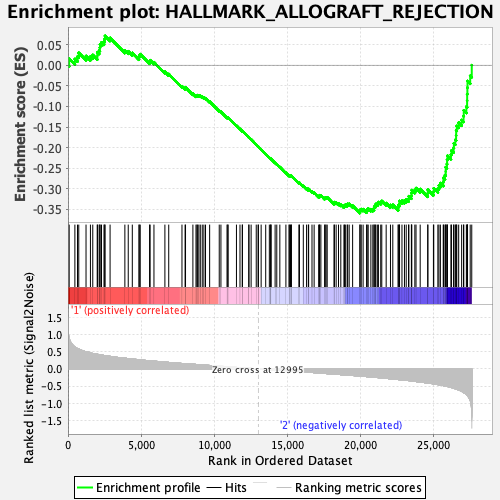
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group2_versus_Group3.MEP.ery_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.35891005 |
| Normalized Enrichment Score (NES) | -1.3378677 |
| Nominal p-value | 0.14202334 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.823 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thy1 | 80 | 0.897 | 0.0159 | No |
| 2 | Tgfb2 | 470 | 0.641 | 0.0151 | No |
| 3 | Irf7 | 649 | 0.590 | 0.0210 | No |
| 4 | Nos2 | 736 | 0.575 | 0.0299 | No |
| 5 | Il4 | 1232 | 0.501 | 0.0224 | No |
| 6 | Stat1 | 1533 | 0.468 | 0.0213 | No |
| 7 | Akt1 | 1693 | 0.447 | 0.0249 | No |
| 8 | Cd79a | 2005 | 0.425 | 0.0224 | No |
| 9 | Rpl3l | 2006 | 0.425 | 0.0314 | No |
| 10 | Prf1 | 2127 | 0.416 | 0.0357 | No |
| 11 | Dyrk3 | 2179 | 0.410 | 0.0424 | No |
| 12 | Tlr3 | 2190 | 0.409 | 0.0506 | No |
| 13 | Ccr2 | 2290 | 0.402 | 0.0554 | No |
| 14 | Eif3a | 2460 | 0.390 | 0.0575 | No |
| 15 | Apbb1 | 2511 | 0.386 | 0.0637 | No |
| 16 | Brca1 | 2530 | 0.386 | 0.0712 | No |
| 17 | Il2ra | 2876 | 0.364 | 0.0662 | No |
| 18 | Cd86 | 3887 | 0.313 | 0.0360 | No |
| 19 | Mmp9 | 4118 | 0.299 | 0.0339 | No |
| 20 | Abce1 | 4395 | 0.286 | 0.0298 | No |
| 21 | Gcnt1 | 4852 | 0.267 | 0.0188 | No |
| 22 | Socs1 | 4869 | 0.266 | 0.0238 | No |
| 23 | Mrpl3 | 4937 | 0.262 | 0.0268 | No |
| 24 | Ube2n | 5576 | 0.235 | 0.0085 | No |
| 25 | Ccnd3 | 5612 | 0.233 | 0.0121 | No |
| 26 | Eif3j1 | 5875 | 0.224 | 0.0072 | No |
| 27 | Ache | 6623 | 0.199 | -0.0158 | No |
| 28 | Cd3d | 6885 | 0.188 | -0.0214 | No |
| 29 | Egfr | 7790 | 0.157 | -0.0510 | No |
| 30 | Bcat1 | 8004 | 0.150 | -0.0557 | No |
| 31 | Nck1 | 8033 | 0.149 | -0.0536 | No |
| 32 | Il2 | 8537 | 0.136 | -0.0691 | No |
| 33 | Cd2 | 8749 | 0.130 | -0.0740 | No |
| 34 | Ptpn6 | 8813 | 0.127 | -0.0737 | No |
| 35 | H2-Aa | 8860 | 0.126 | -0.0727 | No |
| 36 | Eif3d | 8944 | 0.123 | -0.0731 | No |
| 37 | Nlrp3 | 9043 | 0.120 | -0.0742 | No |
| 38 | Jak2 | 9176 | 0.116 | -0.0766 | No |
| 39 | Abi1 | 9289 | 0.112 | -0.0783 | No |
| 40 | Cd4 | 9399 | 0.109 | -0.0800 | No |
| 41 | Timp1 | 9682 | 0.100 | -0.0882 | No |
| 42 | Il2rb | 10338 | 0.080 | -0.1104 | No |
| 43 | Csf1 | 10453 | 0.077 | -0.1129 | No |
| 44 | Npm1 | 10870 | 0.063 | -0.1268 | No |
| 45 | Fcgr2b | 10926 | 0.061 | -0.1275 | No |
| 46 | Tlr6 | 10938 | 0.061 | -0.1266 | No |
| 47 | Wars1 | 11514 | 0.043 | -0.1467 | No |
| 48 | Crtam | 11756 | 0.036 | -0.1547 | No |
| 49 | Traf2 | 11911 | 0.031 | -0.1597 | No |
| 50 | Eif4g3 | 11923 | 0.030 | -0.1595 | No |
| 51 | Tlr1 | 12360 | 0.018 | -0.1750 | No |
| 52 | Ikbkb | 12365 | 0.018 | -0.1748 | No |
| 53 | Psmb10 | 12517 | 0.014 | -0.1800 | No |
| 54 | Map3k7 | 12867 | 0.004 | -0.1926 | No |
| 55 | Cd47 | 12992 | 0.000 | -0.1971 | No |
| 56 | Cd40lg | 13017 | 0.000 | -0.1980 | No |
| 57 | Prkcg | 13202 | -0.001 | -0.2047 | No |
| 58 | Rpl9 | 13514 | -0.011 | -0.2158 | No |
| 59 | Mtif2 | 13762 | -0.018 | -0.2244 | No |
| 60 | Bcl3 | 13850 | -0.020 | -0.2272 | No |
| 61 | Rps3a1 | 13871 | -0.021 | -0.2275 | No |
| 62 | Fgr | 13873 | -0.021 | -0.2271 | No |
| 63 | Acvr2a | 14157 | -0.028 | -0.2368 | No |
| 64 | Tapbp | 14271 | -0.032 | -0.2402 | No |
| 65 | Fasl | 14480 | -0.037 | -0.2470 | No |
| 66 | Il7 | 14894 | -0.050 | -0.2610 | No |
| 67 | Galnt1 | 15110 | -0.056 | -0.2677 | No |
| 68 | Cd3g | 15129 | -0.057 | -0.2672 | No |
| 69 | Irf8 | 15207 | -0.059 | -0.2688 | No |
| 70 | Igsf6 | 15258 | -0.061 | -0.2693 | No |
| 71 | F2 | 15266 | -0.061 | -0.2683 | No |
| 72 | Nme1 | 15783 | -0.076 | -0.2855 | No |
| 73 | Pf4 | 15821 | -0.077 | -0.2852 | No |
| 74 | Rps9 | 16084 | -0.086 | -0.2930 | No |
| 75 | Socs5 | 16307 | -0.092 | -0.2991 | No |
| 76 | Was | 16428 | -0.096 | -0.3015 | No |
| 77 | Gzmb | 16449 | -0.096 | -0.3002 | No |
| 78 | Itk | 16678 | -0.102 | -0.3064 | No |
| 79 | Lck | 16824 | -0.107 | -0.3094 | No |
| 80 | Glmn | 17133 | -0.117 | -0.3182 | No |
| 81 | Rpl39-ps | 17195 | -0.119 | -0.3179 | No |
| 82 | Cd40 | 17197 | -0.120 | -0.3155 | No |
| 83 | Il4ra | 17285 | -0.121 | -0.3161 | No |
| 84 | Il15 | 17541 | -0.130 | -0.3227 | No |
| 85 | Bcl10 | 17579 | -0.131 | -0.3212 | No |
| 86 | Hif1a | 17641 | -0.134 | -0.3207 | No |
| 87 | Cd80 | 17716 | -0.136 | -0.3205 | No |
| 88 | Ube2d1 | 18183 | -0.149 | -0.3344 | No |
| 89 | Lif | 18211 | -0.150 | -0.3322 | No |
| 90 | Ripk2 | 18329 | -0.155 | -0.3332 | No |
| 91 | Stab1 | 18490 | -0.160 | -0.3357 | No |
| 92 | Tlr2 | 18647 | -0.165 | -0.3379 | No |
| 93 | Cd7 | 18863 | -0.171 | -0.3422 | No |
| 94 | Ccnd2 | 18897 | -0.172 | -0.3398 | No |
| 95 | Ccl5 | 18958 | -0.174 | -0.3383 | No |
| 96 | H2-DMb2 | 19080 | -0.179 | -0.3390 | No |
| 97 | Il11 | 19117 | -0.180 | -0.3365 | No |
| 98 | Il10 | 19219 | -0.183 | -0.3364 | No |
| 99 | Cdkn2a | 19455 | -0.190 | -0.3409 | No |
| 100 | B2m | 19949 | -0.208 | -0.3545 | Yes |
| 101 | Csk | 19965 | -0.209 | -0.3507 | Yes |
| 102 | Eif5a | 20044 | -0.211 | -0.3491 | Yes |
| 103 | Ereg | 20172 | -0.216 | -0.3492 | Yes |
| 104 | Gpr65 | 20409 | -0.223 | -0.3532 | Yes |
| 105 | Ccr1 | 20448 | -0.225 | -0.3498 | Yes |
| 106 | Ctss | 20519 | -0.227 | -0.3476 | Yes |
| 107 | Ccr5 | 20702 | -0.231 | -0.3494 | Yes |
| 108 | Tnf | 20837 | -0.237 | -0.3493 | Yes |
| 109 | Hcls1 | 20915 | -0.241 | -0.3471 | Yes |
| 110 | Map4k1 | 20934 | -0.241 | -0.3427 | Yes |
| 111 | Ets1 | 21008 | -0.244 | -0.3402 | Yes |
| 112 | Ly86 | 21048 | -0.247 | -0.3365 | Yes |
| 113 | Tap2 | 21166 | -0.251 | -0.3355 | Yes |
| 114 | Tpd52 | 21218 | -0.253 | -0.3320 | Yes |
| 115 | Ncr1 | 21376 | -0.260 | -0.3323 | Yes |
| 116 | Hdac9 | 21448 | -0.263 | -0.3294 | Yes |
| 117 | Cd3e | 21748 | -0.274 | -0.3345 | Yes |
| 118 | Il1b | 22042 | -0.287 | -0.3392 | Yes |
| 119 | Elane | 22201 | -0.294 | -0.3388 | Yes |
| 120 | Ifnar2 | 22568 | -0.308 | -0.3457 | Yes |
| 121 | Degs1 | 22577 | -0.309 | -0.3395 | Yes |
| 122 | Tap1 | 22656 | -0.312 | -0.3358 | Yes |
| 123 | Cd247 | 22662 | -0.312 | -0.3294 | Yes |
| 124 | Il27ra | 22828 | -0.319 | -0.3287 | Yes |
| 125 | Fas | 22998 | -0.327 | -0.3281 | Yes |
| 126 | Icam1 | 23118 | -0.332 | -0.3254 | Yes |
| 127 | Flna | 23276 | -0.338 | -0.3241 | Yes |
| 128 | Stat4 | 23300 | -0.339 | -0.3178 | Yes |
| 129 | Ifngr2 | 23471 | -0.349 | -0.3167 | Yes |
| 130 | Irf4 | 23480 | -0.350 | -0.3096 | Yes |
| 131 | Il18rap | 23500 | -0.351 | -0.3030 | Yes |
| 132 | Ifngr1 | 23708 | -0.362 | -0.3029 | Yes |
| 133 | Cfp | 23788 | -0.366 | -0.2981 | Yes |
| 134 | H2-Oa | 24073 | -0.381 | -0.3005 | Yes |
| 135 | Icosl | 24592 | -0.414 | -0.3107 | Yes |
| 136 | St8sia4 | 24597 | -0.414 | -0.3022 | Yes |
| 137 | Il18 | 24978 | -0.433 | -0.3069 | Yes |
| 138 | Cd28 | 25001 | -0.435 | -0.2986 | Yes |
| 139 | Prkcb | 25282 | -0.456 | -0.2993 | Yes |
| 140 | F2r | 25354 | -0.461 | -0.2922 | Yes |
| 141 | Ly75 | 25454 | -0.468 | -0.2860 | Yes |
| 142 | Itgal | 25652 | -0.485 | -0.2830 | Yes |
| 143 | Lcp2 | 25662 | -0.486 | -0.2731 | Yes |
| 144 | Ptprc | 25767 | -0.494 | -0.2666 | Yes |
| 145 | H2-DMa | 25817 | -0.500 | -0.2579 | Yes |
| 146 | Ncf4 | 25819 | -0.500 | -0.2474 | Yes |
| 147 | Il2rg | 25900 | -0.507 | -0.2397 | Yes |
| 148 | Spi1 | 25912 | -0.509 | -0.2295 | Yes |
| 149 | Il12a | 25937 | -0.510 | -0.2196 | Yes |
| 150 | Cd1d2 | 26175 | -0.536 | -0.2170 | Yes |
| 151 | Il12rb1 | 26209 | -0.540 | -0.2069 | Yes |
| 152 | Klrd1 | 26356 | -0.560 | -0.2005 | Yes |
| 153 | Il6 | 26377 | -0.563 | -0.1894 | Yes |
| 154 | Srgn | 26475 | -0.577 | -0.1808 | Yes |
| 155 | Trat1 | 26528 | -0.585 | -0.1705 | Yes |
| 156 | Lyn | 26533 | -0.585 | -0.1584 | Yes |
| 157 | Itgb2 | 26574 | -0.591 | -0.1474 | Yes |
| 158 | Tgfb1 | 26704 | -0.611 | -0.1393 | Yes |
| 159 | Inhba | 26907 | -0.651 | -0.1330 | Yes |
| 160 | Il16 | 27036 | -0.682 | -0.1234 | Yes |
| 161 | Elf4 | 27052 | -0.687 | -0.1095 | Yes |
| 162 | Ltb | 27229 | -0.744 | -0.1004 | Yes |
| 163 | Capg | 27281 | -0.768 | -0.0861 | Yes |
| 164 | H2-Ob | 27285 | -0.771 | -0.0701 | Yes |
| 165 | C2 | 27299 | -0.774 | -0.0543 | Yes |
| 166 | Cd74 | 27309 | -0.776 | -0.0384 | Yes |
| 167 | Zap70 | 27492 | -0.921 | -0.0257 | Yes |
| 168 | Cd96 | 27598 | -1.415 | 0.0001 | Yes |