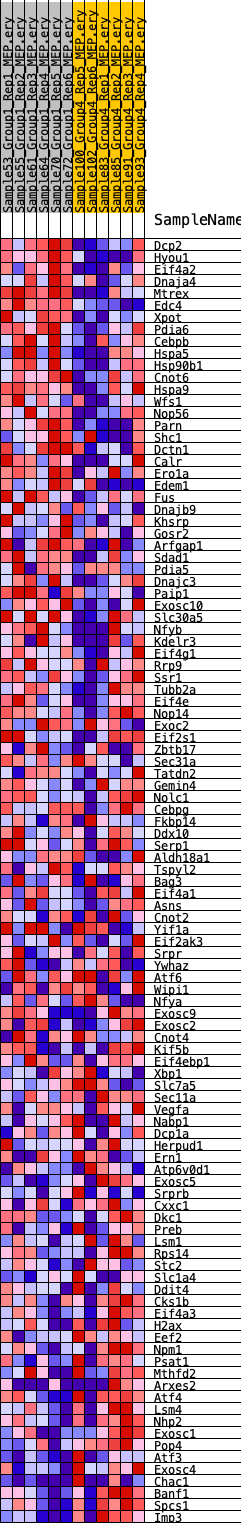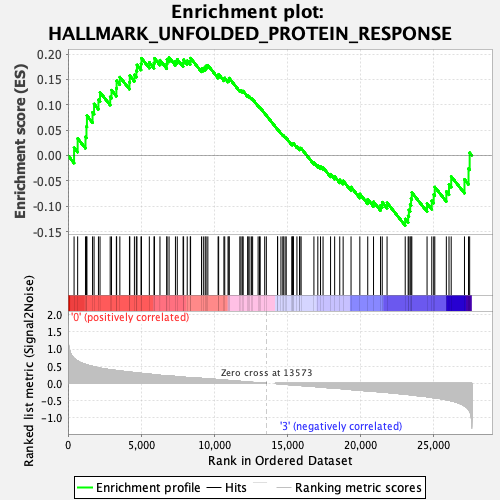
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.19328247 |
| Normalized Enrichment Score (NES) | 0.82177746 |
| Nominal p-value | 0.63809526 |
| FDR q-value | 0.8197342 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcp2 | 415 | 0.726 | 0.0153 | Yes |
| 2 | Hyou1 | 660 | 0.649 | 0.0335 | Yes |
| 3 | Eif4a2 | 1195 | 0.548 | 0.0370 | Yes |
| 4 | Dnaja4 | 1262 | 0.540 | 0.0572 | Yes |
| 5 | Mtrex | 1294 | 0.535 | 0.0784 | Yes |
| 6 | Edc4 | 1678 | 0.493 | 0.0851 | Yes |
| 7 | Xpot | 1782 | 0.484 | 0.1016 | Yes |
| 8 | Pdia6 | 2081 | 0.455 | 0.1098 | Yes |
| 9 | Cebpb | 2189 | 0.444 | 0.1244 | Yes |
| 10 | Hspa5 | 2884 | 0.395 | 0.1157 | Yes |
| 11 | Hsp90b1 | 2974 | 0.389 | 0.1287 | Yes |
| 12 | Cnot6 | 3305 | 0.375 | 0.1324 | Yes |
| 13 | Hspa9 | 3324 | 0.373 | 0.1473 | Yes |
| 14 | Wfs1 | 3544 | 0.362 | 0.1545 | Yes |
| 15 | Nop56 | 4201 | 0.330 | 0.1444 | Yes |
| 16 | Parn | 4225 | 0.329 | 0.1573 | Yes |
| 17 | Shc1 | 4539 | 0.313 | 0.1590 | Yes |
| 18 | Dctn1 | 4671 | 0.309 | 0.1672 | Yes |
| 19 | Calr | 4713 | 0.307 | 0.1785 | Yes |
| 20 | Ero1a | 4992 | 0.293 | 0.1806 | Yes |
| 21 | Edem1 | 5023 | 0.291 | 0.1917 | Yes |
| 22 | Fus | 5559 | 0.268 | 0.1834 | Yes |
| 23 | Dnajb9 | 5878 | 0.252 | 0.1824 | Yes |
| 24 | Khsrp | 5913 | 0.250 | 0.1916 | Yes |
| 25 | Gosr2 | 6286 | 0.234 | 0.1878 | Yes |
| 26 | Arfgap1 | 6752 | 0.225 | 0.1803 | Yes |
| 27 | Sdad1 | 6765 | 0.224 | 0.1893 | Yes |
| 28 | Pdia5 | 6907 | 0.218 | 0.1933 | Yes |
| 29 | Dnajc3 | 7344 | 0.201 | 0.1858 | No |
| 30 | Paip1 | 7469 | 0.195 | 0.1895 | No |
| 31 | Exosc10 | 7867 | 0.180 | 0.1825 | No |
| 32 | Slc30a5 | 7901 | 0.178 | 0.1888 | No |
| 33 | Nfyb | 8149 | 0.169 | 0.1868 | No |
| 34 | Kdelr3 | 8358 | 0.160 | 0.1860 | No |
| 35 | Eif4g1 | 8379 | 0.159 | 0.1919 | No |
| 36 | Rrp9 | 9125 | 0.145 | 0.1708 | No |
| 37 | Ssr1 | 9250 | 0.141 | 0.1722 | No |
| 38 | Tubb2a | 9374 | 0.136 | 0.1734 | No |
| 39 | Eif4e | 9438 | 0.134 | 0.1768 | No |
| 40 | Nop14 | 9559 | 0.130 | 0.1779 | No |
| 41 | Exoc2 | 10261 | 0.108 | 0.1569 | No |
| 42 | Eif2s1 | 10297 | 0.107 | 0.1601 | No |
| 43 | Zbtb17 | 10657 | 0.094 | 0.1510 | No |
| 44 | Sec31a | 10714 | 0.092 | 0.1528 | No |
| 45 | Tatdn2 | 10939 | 0.086 | 0.1482 | No |
| 46 | Gemin4 | 11004 | 0.083 | 0.1494 | No |
| 47 | Nolc1 | 11014 | 0.083 | 0.1525 | No |
| 48 | Cebpg | 11746 | 0.058 | 0.1284 | No |
| 49 | Fkbp14 | 11823 | 0.056 | 0.1280 | No |
| 50 | Ddx10 | 11913 | 0.053 | 0.1269 | No |
| 51 | Serp1 | 11973 | 0.050 | 0.1269 | No |
| 52 | Aldh18a1 | 12281 | 0.040 | 0.1174 | No |
| 53 | Tspyl2 | 12319 | 0.038 | 0.1176 | No |
| 54 | Bag3 | 12431 | 0.035 | 0.1151 | No |
| 55 | Eif4a1 | 12550 | 0.031 | 0.1121 | No |
| 56 | Asns | 12620 | 0.029 | 0.1108 | No |
| 57 | Cnot2 | 12995 | 0.017 | 0.0979 | No |
| 58 | Yif1a | 13089 | 0.016 | 0.0951 | No |
| 59 | Eif2ak3 | 13137 | 0.014 | 0.0940 | No |
| 60 | Srpr | 13445 | 0.004 | 0.0830 | No |
| 61 | Ywhaz | 13557 | 0.000 | 0.0790 | No |
| 62 | Atf6 | 14324 | -0.005 | 0.0513 | No |
| 63 | Wipi1 | 14328 | -0.005 | 0.0515 | No |
| 64 | Nfya | 14555 | -0.013 | 0.0438 | No |
| 65 | Exosc9 | 14695 | -0.018 | 0.0395 | No |
| 66 | Exosc2 | 14705 | -0.018 | 0.0399 | No |
| 67 | Cnot4 | 14836 | -0.024 | 0.0362 | No |
| 68 | Kif5b | 14916 | -0.027 | 0.0344 | No |
| 69 | Eif4ebp1 | 15295 | -0.040 | 0.0223 | No |
| 70 | Xbp1 | 15349 | -0.042 | 0.0222 | No |
| 71 | Slc7a5 | 15379 | -0.043 | 0.0229 | No |
| 72 | Sec11a | 15411 | -0.044 | 0.0237 | No |
| 73 | Vegfa | 15648 | -0.051 | 0.0172 | No |
| 74 | Nabp1 | 15826 | -0.057 | 0.0132 | No |
| 75 | Dcp1a | 15851 | -0.058 | 0.0147 | No |
| 76 | Herpud1 | 15934 | -0.060 | 0.0142 | No |
| 77 | Ern1 | 16809 | -0.086 | -0.0140 | No |
| 78 | Atp6v0d1 | 17076 | -0.096 | -0.0196 | No |
| 79 | Exosc5 | 17261 | -0.103 | -0.0220 | No |
| 80 | Srprb | 17434 | -0.110 | -0.0237 | No |
| 81 | Cxxc1 | 17947 | -0.128 | -0.0369 | No |
| 82 | Dkc1 | 18227 | -0.133 | -0.0415 | No |
| 83 | Preb | 18575 | -0.146 | -0.0481 | No |
| 84 | Lsm1 | 18807 | -0.155 | -0.0500 | No |
| 85 | Rps14 | 19347 | -0.176 | -0.0622 | No |
| 86 | Stc2 | 19948 | -0.196 | -0.0758 | No |
| 87 | Slc1a4 | 20488 | -0.217 | -0.0864 | No |
| 88 | Ddit4 | 20884 | -0.227 | -0.0913 | No |
| 89 | Cks1b | 21363 | -0.246 | -0.0984 | No |
| 90 | Eif4a3 | 21479 | -0.250 | -0.0921 | No |
| 91 | H2ax | 21809 | -0.260 | -0.0931 | No |
| 92 | Eef2 | 23050 | -0.313 | -0.1252 | No |
| 93 | Npm1 | 23251 | -0.321 | -0.1190 | No |
| 94 | Psat1 | 23301 | -0.324 | -0.1073 | No |
| 95 | Mthfd2 | 23388 | -0.328 | -0.0967 | No |
| 96 | Arxes2 | 23449 | -0.330 | -0.0851 | No |
| 97 | Atf4 | 23501 | -0.334 | -0.0730 | No |
| 98 | Lsm4 | 24543 | -0.386 | -0.0947 | No |
| 99 | Nhp2 | 24866 | -0.407 | -0.0894 | No |
| 100 | Exosc1 | 24998 | -0.413 | -0.0769 | No |
| 101 | Pop4 | 25072 | -0.416 | -0.0622 | No |
| 102 | Atf3 | 25861 | -0.473 | -0.0711 | No |
| 103 | Exosc4 | 26043 | -0.491 | -0.0571 | No |
| 104 | Chac1 | 26193 | -0.505 | -0.0414 | No |
| 105 | Banf1 | 27101 | -0.653 | -0.0471 | No |
| 106 | Spcs1 | 27378 | -0.752 | -0.0257 | No |
| 107 | Imp3 | 27455 | -0.809 | 0.0053 | No |