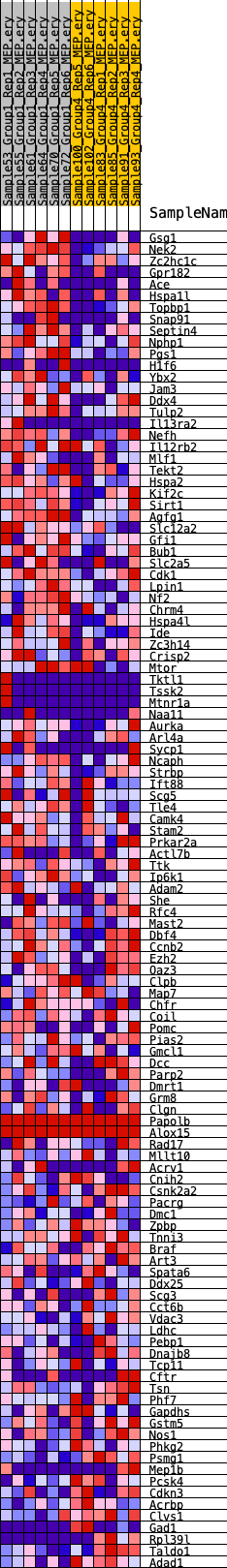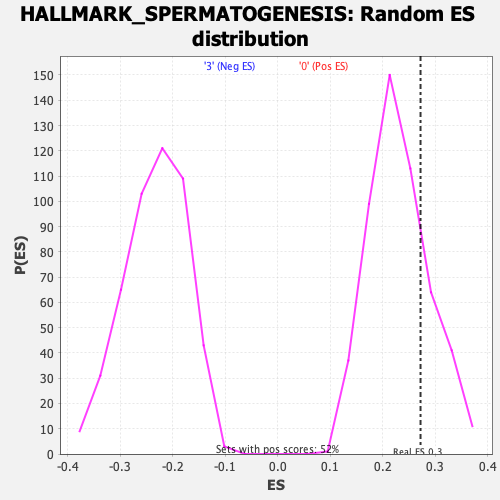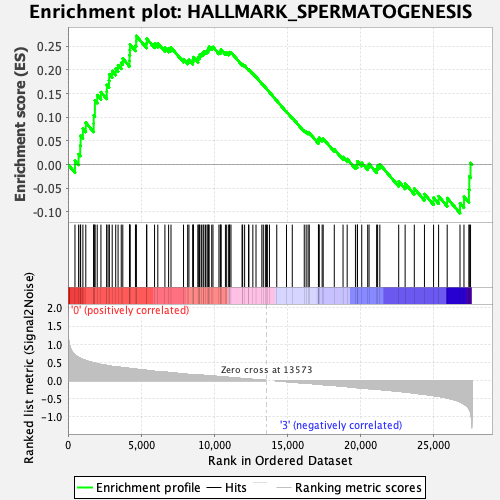
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.2721405 |
| Normalized Enrichment Score (NES) | 1.1782659 |
| Nominal p-value | 0.2248062 |
| FDR q-value | 0.636579 |
| FWER p-Value | 0.965 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gsg1 | 476 | 0.703 | 0.0081 | Yes |
| 2 | Nek2 | 724 | 0.632 | 0.0219 | Yes |
| 3 | Zc2hc1c | 836 | 0.607 | 0.0398 | Yes |
| 4 | Gpr182 | 863 | 0.603 | 0.0607 | Yes |
| 5 | Ace | 1018 | 0.574 | 0.0758 | Yes |
| 6 | Hspa1l | 1216 | 0.546 | 0.0884 | Yes |
| 7 | Topbp1 | 1755 | 0.487 | 0.0864 | Yes |
| 8 | Snap91 | 1758 | 0.487 | 0.1039 | Yes |
| 9 | Septin4 | 1838 | 0.479 | 0.1184 | Yes |
| 10 | Nphp1 | 1839 | 0.479 | 0.1357 | Yes |
| 11 | Pgs1 | 1998 | 0.463 | 0.1466 | Yes |
| 12 | H1f6 | 2251 | 0.436 | 0.1532 | Yes |
| 13 | Ybx2 | 2649 | 0.413 | 0.1537 | Yes |
| 14 | Jam3 | 2655 | 0.413 | 0.1684 | Yes |
| 15 | Ddx4 | 2801 | 0.398 | 0.1775 | Yes |
| 16 | Tulp2 | 2821 | 0.397 | 0.1912 | Yes |
| 17 | Il13ra2 | 3021 | 0.386 | 0.1979 | Yes |
| 18 | Nefh | 3261 | 0.377 | 0.2028 | Yes |
| 19 | Il12rb2 | 3423 | 0.367 | 0.2102 | Yes |
| 20 | Mlf1 | 3639 | 0.357 | 0.2152 | Yes |
| 21 | Tekt2 | 3745 | 0.351 | 0.2241 | Yes |
| 22 | Hspa2 | 4202 | 0.330 | 0.2195 | Yes |
| 23 | Kif2c | 4207 | 0.330 | 0.2312 | Yes |
| 24 | Sirt1 | 4231 | 0.328 | 0.2422 | Yes |
| 25 | Agfg1 | 4241 | 0.327 | 0.2538 | Yes |
| 26 | Slc12a2 | 4611 | 0.312 | 0.2516 | Yes |
| 27 | Gfi1 | 4660 | 0.309 | 0.2610 | Yes |
| 28 | Bub1 | 4663 | 0.309 | 0.2721 | Yes |
| 29 | Slc2a5 | 5372 | 0.279 | 0.2564 | No |
| 30 | Cdk1 | 5384 | 0.278 | 0.2661 | No |
| 31 | Lpin1 | 5916 | 0.250 | 0.2558 | No |
| 32 | Nf2 | 6138 | 0.241 | 0.2564 | No |
| 33 | Chrm4 | 6624 | 0.225 | 0.2469 | No |
| 34 | Hspa4l | 6877 | 0.220 | 0.2457 | No |
| 35 | Ide | 7040 | 0.212 | 0.2475 | No |
| 36 | Zc3h14 | 7896 | 0.178 | 0.2228 | No |
| 37 | Crisp2 | 8181 | 0.167 | 0.2186 | No |
| 38 | Mtor | 8265 | 0.164 | 0.2215 | No |
| 39 | Tktl1 | 8529 | 0.155 | 0.2175 | No |
| 40 | Tssk2 | 8550 | 0.155 | 0.2224 | No |
| 41 | Mtnr1a | 8574 | 0.155 | 0.2272 | No |
| 42 | Naa11 | 8888 | 0.153 | 0.2213 | No |
| 43 | Aurka | 8896 | 0.152 | 0.2265 | No |
| 44 | Arl4a | 8982 | 0.150 | 0.2289 | No |
| 45 | Sycp1 | 9016 | 0.149 | 0.2330 | No |
| 46 | Ncaph | 9133 | 0.144 | 0.2340 | No |
| 47 | Strbp | 9235 | 0.142 | 0.2355 | No |
| 48 | Ift88 | 9288 | 0.139 | 0.2386 | No |
| 49 | Scg5 | 9403 | 0.135 | 0.2394 | No |
| 50 | Tle4 | 9506 | 0.132 | 0.2404 | No |
| 51 | Camk4 | 9575 | 0.130 | 0.2426 | No |
| 52 | Stam2 | 9596 | 0.129 | 0.2466 | No |
| 53 | Prkar2a | 9649 | 0.128 | 0.2493 | No |
| 54 | Actl7b | 9824 | 0.123 | 0.2474 | No |
| 55 | Ttk | 9909 | 0.120 | 0.2487 | No |
| 56 | Ip6k1 | 10318 | 0.107 | 0.2377 | No |
| 57 | Adam2 | 10413 | 0.103 | 0.2380 | No |
| 58 | She | 10445 | 0.102 | 0.2405 | No |
| 59 | Rfc4 | 10462 | 0.101 | 0.2436 | No |
| 60 | Mast2 | 10761 | 0.091 | 0.2361 | No |
| 61 | Dbf4 | 10846 | 0.089 | 0.2362 | No |
| 62 | Ccnb2 | 10982 | 0.084 | 0.2344 | No |
| 63 | Ezh2 | 11034 | 0.082 | 0.2355 | No |
| 64 | Oaz3 | 11053 | 0.082 | 0.2378 | No |
| 65 | Clpb | 11140 | 0.078 | 0.2375 | No |
| 66 | Map7 | 11898 | 0.053 | 0.2118 | No |
| 67 | Chfr | 11937 | 0.052 | 0.2123 | No |
| 68 | Coil | 12073 | 0.047 | 0.2091 | No |
| 69 | Pomc | 12341 | 0.038 | 0.2008 | No |
| 70 | Pias2 | 12364 | 0.037 | 0.2013 | No |
| 71 | Gmcl1 | 12635 | 0.028 | 0.1925 | No |
| 72 | Dcc | 12851 | 0.021 | 0.1854 | No |
| 73 | Parp2 | 13250 | 0.010 | 0.1713 | No |
| 74 | Dmrt1 | 13349 | 0.008 | 0.1680 | No |
| 75 | Grm8 | 13491 | 0.003 | 0.1630 | No |
| 76 | Clgn | 13564 | 0.000 | 0.1604 | No |
| 77 | Papolb | 13628 | 0.000 | 0.1581 | No |
| 78 | Alox15 | 13770 | 0.000 | 0.1530 | No |
| 79 | Rad17 | 14269 | -0.003 | 0.1350 | No |
| 80 | Mllt10 | 14936 | -0.027 | 0.1117 | No |
| 81 | Acrv1 | 15330 | -0.041 | 0.0989 | No |
| 82 | Cnih2 | 16149 | -0.066 | 0.0716 | No |
| 83 | Csnk2a2 | 16278 | -0.070 | 0.0694 | No |
| 84 | Pacrg | 16409 | -0.073 | 0.0673 | No |
| 85 | Dmc1 | 16492 | -0.076 | 0.0671 | No |
| 86 | Zpbp | 17120 | -0.098 | 0.0478 | No |
| 87 | Tnni3 | 17121 | -0.098 | 0.0514 | No |
| 88 | Braf | 17139 | -0.099 | 0.0543 | No |
| 89 | Art3 | 17170 | -0.100 | 0.0568 | No |
| 90 | Spata6 | 17372 | -0.107 | 0.0534 | No |
| 91 | Ddx25 | 17443 | -0.110 | 0.0548 | No |
| 92 | Scg3 | 18201 | -0.132 | 0.0321 | No |
| 93 | Cct6b | 18800 | -0.154 | 0.0159 | No |
| 94 | Vdac3 | 19083 | -0.165 | 0.0116 | No |
| 95 | Ldhc | 19648 | -0.189 | -0.0021 | No |
| 96 | Pebp1 | 19778 | -0.193 | 0.0002 | No |
| 97 | Dnajb8 | 19785 | -0.193 | 0.0069 | No |
| 98 | Tcp11 | 20078 | -0.201 | 0.0036 | No |
| 99 | Cftr | 20486 | -0.217 | -0.0034 | No |
| 100 | Tsn | 20578 | -0.221 | 0.0013 | No |
| 101 | Phf7 | 21096 | -0.235 | -0.0090 | No |
| 102 | Gapdhs | 21163 | -0.238 | -0.0028 | No |
| 103 | Gstm5 | 21315 | -0.244 | 0.0005 | No |
| 104 | Nos1 | 22604 | -0.294 | -0.0357 | No |
| 105 | Phkg2 | 23044 | -0.313 | -0.0404 | No |
| 106 | Psmg1 | 23671 | -0.342 | -0.0508 | No |
| 107 | Mep1b | 24365 | -0.375 | -0.0625 | No |
| 108 | Pcsk4 | 24988 | -0.413 | -0.0702 | No |
| 109 | Cdkn3 | 25332 | -0.433 | -0.0670 | No |
| 110 | Acrbp | 25919 | -0.480 | -0.0710 | No |
| 111 | Clvs1 | 26795 | -0.584 | -0.0817 | No |
| 112 | Gad1 | 27066 | -0.644 | -0.0683 | No |
| 113 | Rpl39l | 27405 | -0.768 | -0.0528 | No |
| 114 | Taldo1 | 27431 | -0.791 | -0.0252 | No |
| 115 | Adad1 | 27515 | -0.868 | 0.0032 | No |