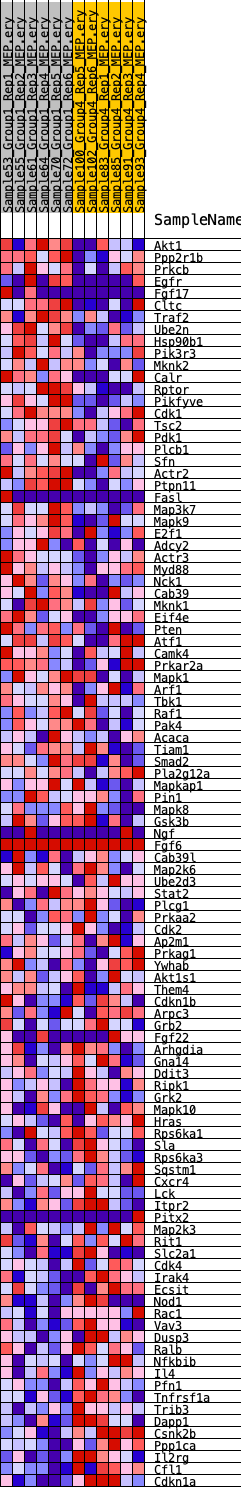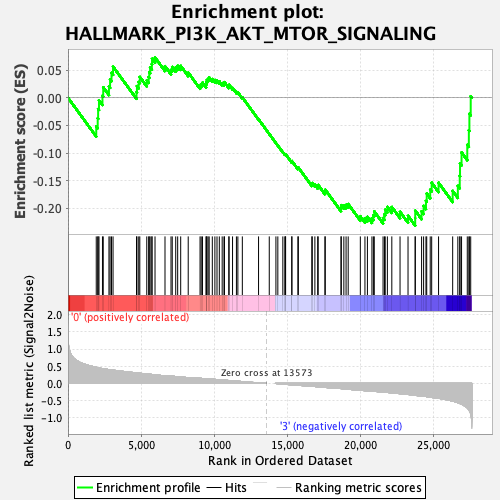
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.23134239 |
| Normalized Enrichment Score (NES) | -1.2254 |
| Nominal p-value | 0.23608445 |
| FDR q-value | 0.49147767 |
| FWER p-Value | 0.945 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 1931 | 0.469 | -0.0518 | No |
| 2 | Ppp2r1b | 2031 | 0.460 | -0.0372 | No |
| 3 | Prkcb | 2060 | 0.458 | -0.0202 | No |
| 4 | Egfr | 2118 | 0.451 | -0.0045 | No |
| 5 | Fgf17 | 2356 | 0.432 | 0.0039 | No |
| 6 | Cltc | 2408 | 0.430 | 0.0190 | No |
| 7 | Traf2 | 2800 | 0.398 | 0.0204 | No |
| 8 | Ube2n | 2876 | 0.396 | 0.0333 | No |
| 9 | Hsp90b1 | 2974 | 0.389 | 0.0451 | No |
| 10 | Pik3r3 | 3079 | 0.383 | 0.0564 | No |
| 11 | Mknk2 | 4682 | 0.308 | 0.0103 | No |
| 12 | Calr | 4713 | 0.307 | 0.0213 | No |
| 13 | Rptor | 4836 | 0.301 | 0.0287 | No |
| 14 | Pikfyve | 4908 | 0.296 | 0.0377 | No |
| 15 | Cdk1 | 5384 | 0.278 | 0.0314 | No |
| 16 | Tsc2 | 5513 | 0.270 | 0.0374 | No |
| 17 | Pdk1 | 5555 | 0.268 | 0.0465 | No |
| 18 | Plcb1 | 5614 | 0.265 | 0.0548 | No |
| 19 | Sfn | 5731 | 0.260 | 0.0608 | No |
| 20 | Actr2 | 5745 | 0.259 | 0.0706 | No |
| 21 | Ptpn11 | 5948 | 0.249 | 0.0730 | No |
| 22 | Fasl | 6630 | 0.225 | 0.0571 | No |
| 23 | Map3k7 | 7041 | 0.212 | 0.0506 | No |
| 24 | Mapk9 | 7125 | 0.208 | 0.0558 | No |
| 25 | E2f1 | 7363 | 0.200 | 0.0550 | No |
| 26 | Adcy2 | 7497 | 0.194 | 0.0578 | No |
| 27 | Actr3 | 7703 | 0.186 | 0.0577 | No |
| 28 | Myd88 | 8217 | 0.166 | 0.0456 | No |
| 29 | Nck1 | 9026 | 0.149 | 0.0221 | No |
| 30 | Cab39 | 9105 | 0.145 | 0.0249 | No |
| 31 | Mknk1 | 9192 | 0.143 | 0.0274 | No |
| 32 | Eif4e | 9438 | 0.134 | 0.0238 | No |
| 33 | Pten | 9453 | 0.134 | 0.0286 | No |
| 34 | Atf1 | 9496 | 0.132 | 0.0323 | No |
| 35 | Camk4 | 9575 | 0.130 | 0.0345 | No |
| 36 | Prkar2a | 9649 | 0.128 | 0.0369 | No |
| 37 | Mapk1 | 9864 | 0.121 | 0.0339 | No |
| 38 | Arf1 | 10040 | 0.116 | 0.0321 | No |
| 39 | Tbk1 | 10185 | 0.111 | 0.0312 | No |
| 40 | Raf1 | 10343 | 0.106 | 0.0297 | No |
| 41 | Pak4 | 10538 | 0.099 | 0.0265 | No |
| 42 | Acaca | 10662 | 0.094 | 0.0258 | No |
| 43 | Tiam1 | 10695 | 0.093 | 0.0283 | No |
| 44 | Smad2 | 10995 | 0.084 | 0.0207 | No |
| 45 | Pla2g12a | 11007 | 0.083 | 0.0236 | No |
| 46 | Mapkap1 | 11247 | 0.074 | 0.0178 | No |
| 47 | Pin1 | 11512 | 0.066 | 0.0108 | No |
| 48 | Mapk8 | 11611 | 0.062 | 0.0097 | No |
| 49 | Gsk3b | 11916 | 0.052 | 0.0007 | No |
| 50 | Ngf | 13025 | 0.016 | -0.0390 | No |
| 51 | Fgf6 | 13757 | 0.000 | -0.0656 | No |
| 52 | Cab39l | 14212 | -0.002 | -0.0820 | No |
| 53 | Map2k6 | 14343 | -0.006 | -0.0865 | No |
| 54 | Ube2d3 | 14685 | -0.017 | -0.0982 | No |
| 55 | Stat2 | 14822 | -0.023 | -0.1023 | No |
| 56 | Plcg1 | 14856 | -0.024 | -0.1025 | No |
| 57 | Prkaa2 | 14875 | -0.025 | -0.1022 | No |
| 58 | Cdk2 | 15298 | -0.040 | -0.1159 | No |
| 59 | Ap2m1 | 15303 | -0.041 | -0.1145 | No |
| 60 | Prkag1 | 15715 | -0.053 | -0.1273 | No |
| 61 | Ywhab | 15744 | -0.054 | -0.1262 | No |
| 62 | Akt1s1 | 16667 | -0.081 | -0.1566 | No |
| 63 | Them4 | 16695 | -0.082 | -0.1543 | No |
| 64 | Cdkn1b | 16875 | -0.089 | -0.1573 | No |
| 65 | Arpc3 | 17068 | -0.096 | -0.1605 | No |
| 66 | Grb2 | 17099 | -0.097 | -0.1578 | No |
| 67 | Fgf22 | 17562 | -0.114 | -0.1701 | No |
| 68 | Arhgdia | 17583 | -0.115 | -0.1663 | No |
| 69 | Gna14 | 18659 | -0.149 | -0.1995 | No |
| 70 | Ddit3 | 18680 | -0.149 | -0.1944 | No |
| 71 | Ripk1 | 18855 | -0.156 | -0.1946 | No |
| 72 | Grk2 | 19007 | -0.162 | -0.1937 | No |
| 73 | Mapk10 | 19152 | -0.168 | -0.1923 | No |
| 74 | Hras | 19982 | -0.198 | -0.2147 | No |
| 75 | Rps6ka1 | 20297 | -0.210 | -0.2178 | No |
| 76 | Sla | 20466 | -0.216 | -0.2154 | No |
| 77 | Rps6ka3 | 20775 | -0.222 | -0.2179 | Yes |
| 78 | Sqstm1 | 20875 | -0.226 | -0.2126 | Yes |
| 79 | Cxcr4 | 20935 | -0.229 | -0.2057 | Yes |
| 80 | Lck | 21535 | -0.253 | -0.2175 | Yes |
| 81 | Itpr2 | 21632 | -0.258 | -0.2109 | Yes |
| 82 | Pitx2 | 21682 | -0.260 | -0.2024 | Yes |
| 83 | Map2k3 | 21830 | -0.261 | -0.1975 | Yes |
| 84 | Rit1 | 22134 | -0.275 | -0.1977 | Yes |
| 85 | Slc2a1 | 22696 | -0.297 | -0.2063 | Yes |
| 86 | Cdk4 | 23242 | -0.320 | -0.2136 | Yes |
| 87 | Irak4 | 23732 | -0.345 | -0.2177 | Yes |
| 88 | Ecsit | 23735 | -0.346 | -0.2042 | Yes |
| 89 | Nod1 | 24169 | -0.367 | -0.2055 | Yes |
| 90 | Rac1 | 24307 | -0.373 | -0.1957 | Yes |
| 91 | Vav3 | 24468 | -0.381 | -0.1866 | Yes |
| 92 | Dusp3 | 24520 | -0.384 | -0.1733 | Yes |
| 93 | Ralb | 24759 | -0.400 | -0.1662 | Yes |
| 94 | Nfkbib | 24859 | -0.407 | -0.1538 | Yes |
| 95 | Il4 | 25329 | -0.432 | -0.1538 | Yes |
| 96 | Pfn1 | 26294 | -0.514 | -0.1686 | Yes |
| 97 | Tnfrsf1a | 26647 | -0.559 | -0.1594 | Yes |
| 98 | Trib3 | 26777 | -0.579 | -0.1413 | Yes |
| 99 | Dapp1 | 26797 | -0.585 | -0.1189 | Yes |
| 100 | Csnk2b | 26901 | -0.604 | -0.0989 | Yes |
| 101 | Ppp1ca | 27292 | -0.710 | -0.0851 | Yes |
| 102 | Il2rg | 27400 | -0.765 | -0.0589 | Yes |
| 103 | Cfl1 | 27437 | -0.794 | -0.0289 | Yes |
| 104 | Cdkn1a | 27527 | -0.885 | 0.0027 | Yes |