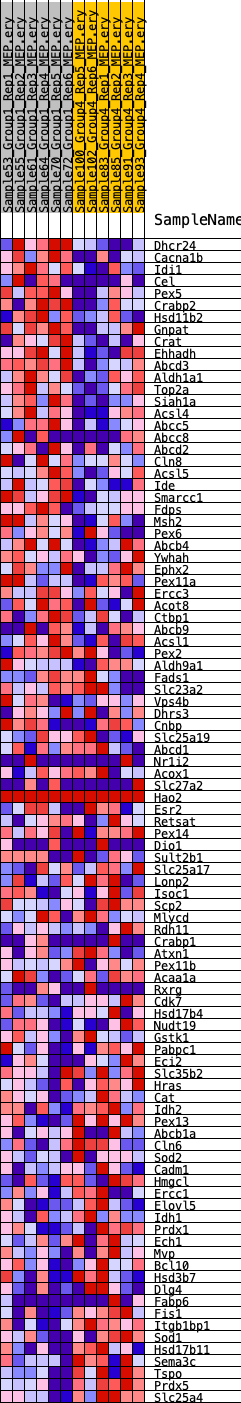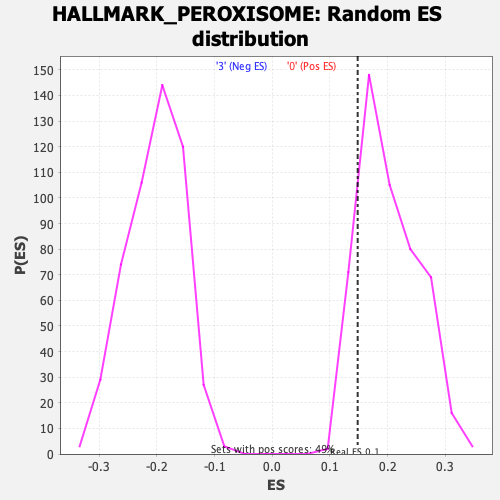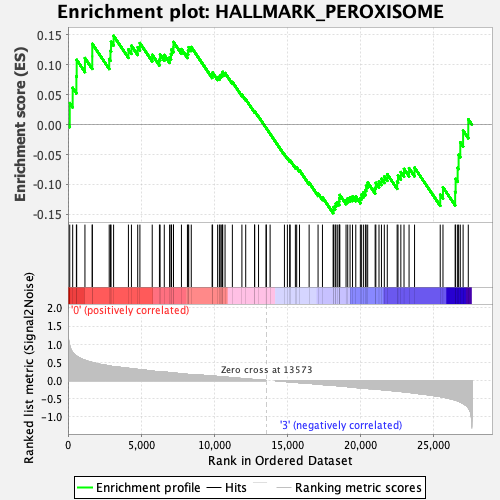
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.1482225 |
| Normalized Enrichment Score (NES) | 0.73336303 |
| Nominal p-value | 0.8582996 |
| FDR q-value | 0.882139 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dhcr24 | 119 | 0.950 | 0.0359 | Yes |
| 2 | Cacna1b | 320 | 0.772 | 0.0613 | Yes |
| 3 | Idi1 | 573 | 0.672 | 0.0806 | Yes |
| 4 | Cel | 594 | 0.666 | 0.1081 | Yes |
| 5 | Pex5 | 1158 | 0.555 | 0.1112 | Yes |
| 6 | Crabp2 | 1658 | 0.495 | 0.1140 | Yes |
| 7 | Hsd11b2 | 1659 | 0.495 | 0.1349 | Yes |
| 8 | Gnpat | 2817 | 0.397 | 0.1096 | Yes |
| 9 | Crat | 2901 | 0.394 | 0.1233 | Yes |
| 10 | Ehhadh | 2942 | 0.391 | 0.1384 | Yes |
| 11 | Abcd3 | 3117 | 0.381 | 0.1482 | Yes |
| 12 | Aldh1a1 | 4130 | 0.335 | 0.1256 | No |
| 13 | Top2a | 4336 | 0.322 | 0.1318 | No |
| 14 | Siah1a | 4765 | 0.304 | 0.1291 | No |
| 15 | Acsl4 | 4915 | 0.296 | 0.1362 | No |
| 16 | Abcc5 | 5757 | 0.259 | 0.1166 | No |
| 17 | Abcc8 | 6250 | 0.236 | 0.1087 | No |
| 18 | Abcd2 | 6294 | 0.234 | 0.1170 | No |
| 19 | Cln8 | 6581 | 0.228 | 0.1163 | No |
| 20 | Acsl5 | 6944 | 0.217 | 0.1123 | No |
| 21 | Ide | 7040 | 0.212 | 0.1178 | No |
| 22 | Smarcc1 | 7074 | 0.211 | 0.1256 | No |
| 23 | Fdps | 7208 | 0.205 | 0.1294 | No |
| 24 | Msh2 | 7210 | 0.205 | 0.1380 | No |
| 25 | Pex6 | 7754 | 0.184 | 0.1261 | No |
| 26 | Abcb4 | 8163 | 0.168 | 0.1184 | No |
| 27 | Ywhah | 8209 | 0.166 | 0.1238 | No |
| 28 | Ephx2 | 8251 | 0.164 | 0.1292 | No |
| 29 | Pex11a | 8426 | 0.157 | 0.1296 | No |
| 30 | Ercc3 | 9851 | 0.122 | 0.0830 | No |
| 31 | Acot8 | 9882 | 0.121 | 0.0870 | No |
| 32 | Ctbp1 | 10228 | 0.109 | 0.0791 | No |
| 33 | Abcb9 | 10346 | 0.105 | 0.0793 | No |
| 34 | Acsl1 | 10393 | 0.104 | 0.0820 | No |
| 35 | Pex2 | 10475 | 0.101 | 0.0833 | No |
| 36 | Aldh9a1 | 10557 | 0.098 | 0.0845 | No |
| 37 | Fads1 | 10576 | 0.097 | 0.0880 | No |
| 38 | Slc23a2 | 10735 | 0.092 | 0.0861 | No |
| 39 | Vps4b | 11235 | 0.075 | 0.0712 | No |
| 40 | Dhrs3 | 11887 | 0.054 | 0.0498 | No |
| 41 | Cnbp | 12147 | 0.044 | 0.0422 | No |
| 42 | Slc25a19 | 12753 | 0.024 | 0.0213 | No |
| 43 | Abcd1 | 12759 | 0.024 | 0.0221 | No |
| 44 | Nr1i2 | 13024 | 0.016 | 0.0132 | No |
| 45 | Acox1 | 13536 | 0.001 | -0.0053 | No |
| 46 | Slc27a2 | 13559 | 0.000 | -0.0061 | No |
| 47 | Hao2 | 13815 | 0.000 | -0.0154 | No |
| 48 | Esr2 | 14794 | -0.022 | -0.0500 | No |
| 49 | Retsat | 14996 | -0.030 | -0.0561 | No |
| 50 | Pex14 | 15153 | -0.035 | -0.0603 | No |
| 51 | Dio1 | 15184 | -0.036 | -0.0598 | No |
| 52 | Sult2b1 | 15542 | -0.049 | -0.0708 | No |
| 53 | Slc25a17 | 15631 | -0.051 | -0.0718 | No |
| 54 | Lonp2 | 15825 | -0.057 | -0.0764 | No |
| 55 | Isoc1 | 16482 | -0.075 | -0.0971 | No |
| 56 | Scp2 | 17093 | -0.097 | -0.1152 | No |
| 57 | Mlycd | 17393 | -0.108 | -0.1215 | No |
| 58 | Rdh11 | 18123 | -0.129 | -0.1425 | No |
| 59 | Crabp1 | 18153 | -0.131 | -0.1380 | No |
| 60 | Atxn1 | 18259 | -0.134 | -0.1362 | No |
| 61 | Pex11b | 18303 | -0.135 | -0.1320 | No |
| 62 | Acaa1a | 18405 | -0.140 | -0.1298 | No |
| 63 | Rxrg | 18539 | -0.145 | -0.1285 | No |
| 64 | Cdk7 | 18544 | -0.145 | -0.1225 | No |
| 65 | Hsd17b4 | 18581 | -0.146 | -0.1176 | No |
| 66 | Nudt19 | 19006 | -0.162 | -0.1262 | No |
| 67 | Gstk1 | 19120 | -0.166 | -0.1233 | No |
| 68 | Pabpc1 | 19280 | -0.173 | -0.1217 | No |
| 69 | Eci2 | 19448 | -0.181 | -0.1201 | No |
| 70 | Slc35b2 | 19671 | -0.189 | -0.1202 | No |
| 71 | Hras | 19982 | -0.198 | -0.1231 | No |
| 72 | Cat | 20051 | -0.200 | -0.1171 | No |
| 73 | Idh2 | 20190 | -0.206 | -0.1133 | No |
| 74 | Pex13 | 20328 | -0.211 | -0.1094 | No |
| 75 | Abcb1a | 20377 | -0.212 | -0.1021 | No |
| 76 | Cln6 | 20485 | -0.217 | -0.0969 | No |
| 77 | Sod2 | 20995 | -0.231 | -0.1056 | No |
| 78 | Cadm1 | 21042 | -0.233 | -0.0974 | No |
| 79 | Hmgcl | 21258 | -0.241 | -0.0950 | No |
| 80 | Ercc1 | 21426 | -0.248 | -0.0905 | No |
| 81 | Elovl5 | 21621 | -0.257 | -0.0867 | No |
| 82 | Idh1 | 21824 | -0.261 | -0.0830 | No |
| 83 | Prdx1 | 22498 | -0.288 | -0.0953 | No |
| 84 | Ech1 | 22557 | -0.292 | -0.0850 | No |
| 85 | Mvp | 22751 | -0.299 | -0.0794 | No |
| 86 | Bcl10 | 22968 | -0.310 | -0.0741 | No |
| 87 | Hsd3b7 | 23313 | -0.324 | -0.0729 | No |
| 88 | Dlg4 | 23688 | -0.343 | -0.0720 | No |
| 89 | Fabp6 | 25443 | -0.441 | -0.1171 | No |
| 90 | Fis1 | 25631 | -0.452 | -0.1047 | No |
| 91 | Itgb1bp1 | 26463 | -0.533 | -0.1124 | No |
| 92 | Sod1 | 26496 | -0.538 | -0.0907 | No |
| 93 | Hsd17b11 | 26634 | -0.556 | -0.0722 | No |
| 94 | Sema3c | 26700 | -0.568 | -0.0505 | No |
| 95 | Tspo | 26814 | -0.588 | -0.0297 | No |
| 96 | Prdx5 | 27007 | -0.630 | -0.0100 | No |
| 97 | Slc25a4 | 27361 | -0.746 | 0.0088 | No |