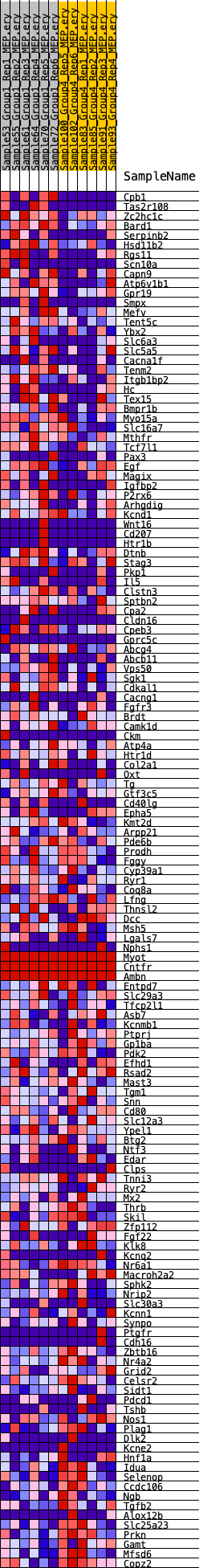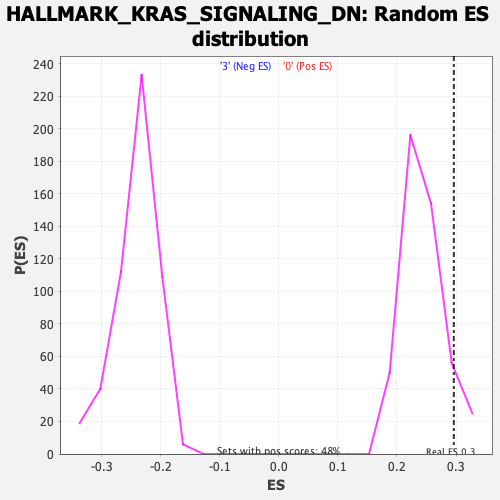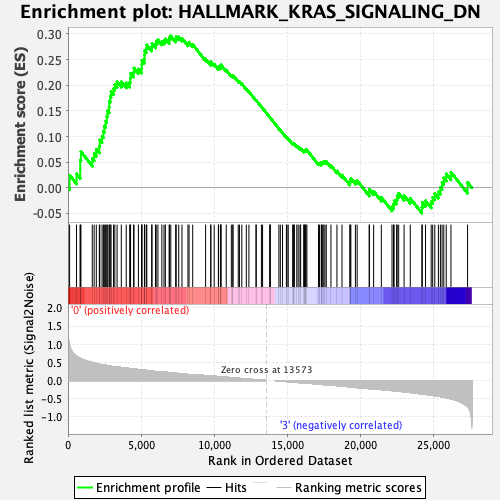
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.29626423 |
| Normalized Enrichment Score (NES) | 1.2214025 |
| Nominal p-value | 0.07900208 |
| FDR q-value | 0.7979357 |
| FWER p-Value | 0.946 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cpb1 | 109 | 0.960 | 0.0246 | Yes |
| 2 | Tas2r108 | 583 | 0.669 | 0.0273 | Yes |
| 3 | Zc2hc1c | 836 | 0.607 | 0.0362 | Yes |
| 4 | Bard1 | 838 | 0.607 | 0.0543 | Yes |
| 5 | Serpinb2 | 884 | 0.598 | 0.0704 | Yes |
| 6 | Hsd11b2 | 1659 | 0.495 | 0.0569 | Yes |
| 7 | Rgs11 | 1787 | 0.484 | 0.0667 | Yes |
| 8 | Scn10a | 1935 | 0.468 | 0.0753 | Yes |
| 9 | Capn9 | 2140 | 0.449 | 0.0812 | Yes |
| 10 | Atp6v1b1 | 2175 | 0.445 | 0.0933 | Yes |
| 11 | Gpr19 | 2328 | 0.433 | 0.1006 | Yes |
| 12 | Smpx | 2420 | 0.429 | 0.1101 | Yes |
| 13 | Mefv | 2485 | 0.426 | 0.1204 | Yes |
| 14 | Tent5c | 2571 | 0.420 | 0.1298 | Yes |
| 15 | Ybx2 | 2649 | 0.413 | 0.1393 | Yes |
| 16 | Slc6a3 | 2686 | 0.409 | 0.1502 | Yes |
| 17 | Slc5a5 | 2809 | 0.397 | 0.1576 | Yes |
| 18 | Cacna1f | 2818 | 0.397 | 0.1691 | Yes |
| 19 | Tenm2 | 2889 | 0.395 | 0.1783 | Yes |
| 20 | Itgb1bp2 | 2944 | 0.391 | 0.1880 | Yes |
| 21 | Hc | 3112 | 0.381 | 0.1932 | Yes |
| 22 | Tex15 | 3201 | 0.378 | 0.2013 | Yes |
| 23 | Bmpr1b | 3355 | 0.370 | 0.2068 | Yes |
| 24 | Myo15a | 3643 | 0.357 | 0.2069 | Yes |
| 25 | Slc16a7 | 3980 | 0.336 | 0.2047 | Yes |
| 26 | Mthfr | 4219 | 0.329 | 0.2058 | Yes |
| 27 | Tcf7l1 | 4257 | 0.327 | 0.2142 | Yes |
| 28 | Pax3 | 4278 | 0.326 | 0.2231 | Yes |
| 29 | Egf | 4484 | 0.316 | 0.2251 | Yes |
| 30 | Magix | 4510 | 0.315 | 0.2336 | Yes |
| 31 | Igfbp2 | 4812 | 0.302 | 0.2316 | Yes |
| 32 | P2rx6 | 5033 | 0.290 | 0.2322 | Yes |
| 33 | Arhgdig | 5044 | 0.290 | 0.2405 | Yes |
| 34 | Kcnd1 | 5052 | 0.290 | 0.2489 | Yes |
| 35 | Wnt16 | 5225 | 0.281 | 0.2510 | Yes |
| 36 | Cd207 | 5229 | 0.281 | 0.2592 | Yes |
| 37 | Htr1b | 5240 | 0.281 | 0.2673 | Yes |
| 38 | Dtnb | 5350 | 0.280 | 0.2716 | Yes |
| 39 | Stag3 | 5388 | 0.277 | 0.2785 | Yes |
| 40 | Pkp1 | 5722 | 0.261 | 0.2742 | Yes |
| 41 | Il5 | 5741 | 0.260 | 0.2813 | Yes |
| 42 | Clstn3 | 5979 | 0.248 | 0.2800 | Yes |
| 43 | Sptbn2 | 6026 | 0.246 | 0.2856 | Yes |
| 44 | Cpa2 | 6146 | 0.240 | 0.2885 | Yes |
| 45 | Cldn16 | 6415 | 0.229 | 0.2855 | Yes |
| 46 | Cpeb3 | 6563 | 0.228 | 0.2870 | Yes |
| 47 | Gprc5c | 6655 | 0.225 | 0.2903 | Yes |
| 48 | Abcg4 | 6917 | 0.218 | 0.2873 | Yes |
| 49 | Abcb11 | 6931 | 0.217 | 0.2933 | Yes |
| 50 | Vps50 | 7026 | 0.214 | 0.2963 | Yes |
| 51 | Sgk1 | 7361 | 0.200 | 0.2901 | No |
| 52 | Cdkal1 | 7394 | 0.198 | 0.2948 | No |
| 53 | Cacng1 | 7568 | 0.191 | 0.2942 | No |
| 54 | Fgfr3 | 7791 | 0.182 | 0.2915 | No |
| 55 | Brdt | 8200 | 0.167 | 0.2816 | No |
| 56 | Camk1d | 8274 | 0.164 | 0.2838 | No |
| 57 | Ckm | 8524 | 0.155 | 0.2794 | No |
| 58 | Atp4a | 9406 | 0.135 | 0.2513 | No |
| 59 | Htr1d | 9757 | 0.125 | 0.2423 | No |
| 60 | Col2a1 | 9763 | 0.124 | 0.2458 | No |
| 61 | Oxt | 9992 | 0.117 | 0.2410 | No |
| 62 | Tg | 10278 | 0.108 | 0.2338 | No |
| 63 | Gtf3c5 | 10293 | 0.107 | 0.2365 | No |
| 64 | Cd40lg | 10424 | 0.103 | 0.2348 | No |
| 65 | Epha5 | 10451 | 0.102 | 0.2369 | No |
| 66 | Kmt2d | 10456 | 0.102 | 0.2398 | No |
| 67 | Arpp21 | 10816 | 0.089 | 0.2294 | No |
| 68 | Pde6b | 11164 | 0.077 | 0.2190 | No |
| 69 | Prodh | 11272 | 0.073 | 0.2173 | No |
| 70 | Fggy | 11273 | 0.073 | 0.2195 | No |
| 71 | Cyp39a1 | 11636 | 0.061 | 0.2081 | No |
| 72 | Ryr1 | 11719 | 0.059 | 0.2069 | No |
| 73 | Coq8a | 11880 | 0.054 | 0.2027 | No |
| 74 | Lfng | 12187 | 0.043 | 0.1928 | No |
| 75 | Thnsl2 | 12366 | 0.037 | 0.1874 | No |
| 76 | Dcc | 12851 | 0.021 | 0.1704 | No |
| 77 | Msh5 | 12866 | 0.021 | 0.1705 | No |
| 78 | Lgals7 | 13220 | 0.011 | 0.1580 | No |
| 79 | Nphs1 | 13300 | 0.008 | 0.1554 | No |
| 80 | Myot | 13790 | 0.000 | 0.1376 | No |
| 81 | Cntfr | 13818 | 0.000 | 0.1366 | No |
| 82 | Ambn | 13822 | 0.000 | 0.1365 | No |
| 83 | Entpd7 | 14416 | -0.008 | 0.1151 | No |
| 84 | Slc29a3 | 14514 | -0.012 | 0.1120 | No |
| 85 | Tfcp2l1 | 14664 | -0.017 | 0.1070 | No |
| 86 | Asb7 | 14927 | -0.027 | 0.0983 | No |
| 87 | Kcnmb1 | 14971 | -0.029 | 0.0976 | No |
| 88 | Ptprj | 15054 | -0.031 | 0.0955 | No |
| 89 | Gp1ba | 15367 | -0.043 | 0.0854 | No |
| 90 | Pdk2 | 15384 | -0.044 | 0.0861 | No |
| 91 | Efhd1 | 15398 | -0.044 | 0.0870 | No |
| 92 | Rsad2 | 15477 | -0.046 | 0.0855 | No |
| 93 | Mast3 | 15639 | -0.051 | 0.0812 | No |
| 94 | Tgm1 | 15732 | -0.054 | 0.0794 | No |
| 95 | Snn | 15857 | -0.058 | 0.0766 | No |
| 96 | Cd80 | 15903 | -0.059 | 0.0767 | No |
| 97 | Slc12a3 | 16113 | -0.065 | 0.0711 | No |
| 98 | Ypel1 | 16143 | -0.066 | 0.0720 | No |
| 99 | Btg2 | 16173 | -0.067 | 0.0729 | No |
| 100 | Ntf3 | 16248 | -0.069 | 0.0723 | No |
| 101 | Edar | 16253 | -0.069 | 0.0742 | No |
| 102 | Clps | 16329 | -0.071 | 0.0736 | No |
| 103 | Tnni3 | 17121 | -0.098 | 0.0477 | No |
| 104 | Ryr2 | 17194 | -0.101 | 0.0481 | No |
| 105 | Mx2 | 17315 | -0.105 | 0.0468 | No |
| 106 | Thrb | 17325 | -0.106 | 0.0496 | No |
| 107 | Skil | 17407 | -0.109 | 0.0499 | No |
| 108 | Zfp112 | 17484 | -0.111 | 0.0505 | No |
| 109 | Fgf22 | 17562 | -0.114 | 0.0511 | No |
| 110 | Klk8 | 17653 | -0.118 | 0.0513 | No |
| 111 | Kcnq2 | 17973 | -0.129 | 0.0435 | No |
| 112 | Nr6a1 | 18383 | -0.139 | 0.0327 | No |
| 113 | Macroh2a2 | 18729 | -0.152 | 0.0247 | No |
| 114 | Sphk2 | 19266 | -0.173 | 0.0103 | No |
| 115 | Nrip2 | 19286 | -0.174 | 0.0148 | No |
| 116 | Slc30a3 | 19334 | -0.176 | 0.0183 | No |
| 117 | Kcnn1 | 19645 | -0.188 | 0.0126 | No |
| 118 | Synpo | 19765 | -0.193 | 0.0140 | No |
| 119 | Ptgfr | 20592 | -0.221 | -0.0095 | No |
| 120 | Cdh16 | 20595 | -0.221 | -0.0029 | No |
| 121 | Zbtb16 | 20892 | -0.227 | -0.0070 | No |
| 122 | Nr4a2 | 21417 | -0.248 | -0.0187 | No |
| 123 | Grid2 | 22147 | -0.276 | -0.0370 | No |
| 124 | Celsr2 | 22247 | -0.281 | -0.0322 | No |
| 125 | Sidt1 | 22291 | -0.281 | -0.0255 | No |
| 126 | Pdcd1 | 22462 | -0.287 | -0.0231 | No |
| 127 | Tshb | 22500 | -0.288 | -0.0159 | No |
| 128 | Nos1 | 22604 | -0.294 | -0.0109 | No |
| 129 | Plag1 | 22972 | -0.310 | -0.0150 | No |
| 130 | Dlk2 | 23396 | -0.328 | -0.0206 | No |
| 131 | Kcne2 | 24190 | -0.368 | -0.0386 | No |
| 132 | Hnf1a | 24215 | -0.368 | -0.0285 | No |
| 133 | Idua | 24438 | -0.379 | -0.0253 | No |
| 134 | Selenop | 24825 | -0.405 | -0.0273 | No |
| 135 | Ccdc106 | 24928 | -0.411 | -0.0187 | No |
| 136 | Ngb | 25071 | -0.416 | -0.0115 | No |
| 137 | Tgfb2 | 25318 | -0.432 | -0.0076 | No |
| 138 | Alox12b | 25455 | -0.441 | 0.0005 | No |
| 139 | Slc25a23 | 25569 | -0.449 | 0.0098 | No |
| 140 | Prkn | 25686 | -0.457 | 0.0192 | No |
| 141 | Gamt | 25852 | -0.472 | 0.0272 | No |
| 142 | Mfsd6 | 26177 | -0.504 | 0.0304 | No |
| 143 | Copz2 | 27312 | -0.720 | 0.0106 | No |