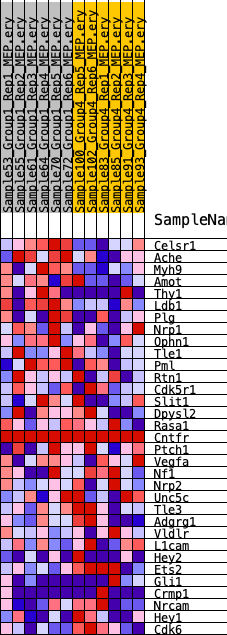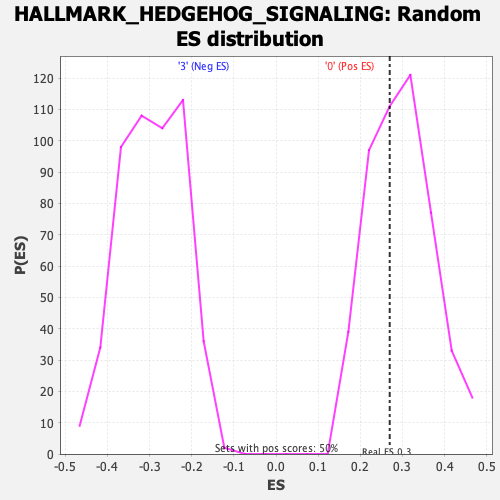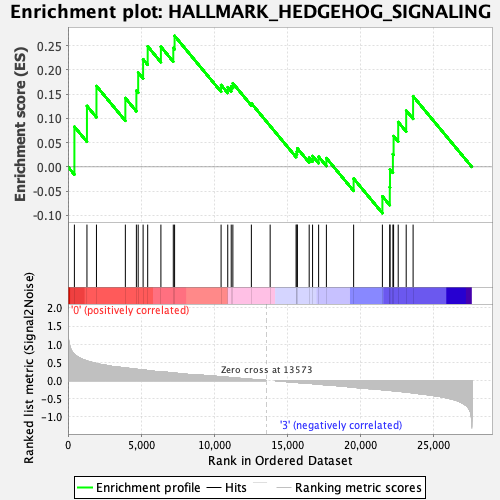
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.26993614 |
| Normalized Enrichment Score (NES) | 0.9080618 |
| Nominal p-value | 0.6028226 |
| FDR q-value | 0.8498047 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Celsr1 | 437 | 0.715 | 0.0828 | Yes |
| 2 | Ache | 1295 | 0.535 | 0.1256 | Yes |
| 3 | Myh9 | 1945 | 0.467 | 0.1665 | Yes |
| 4 | Amot | 3920 | 0.340 | 0.1418 | Yes |
| 5 | Thy1 | 4674 | 0.309 | 0.1571 | Yes |
| 6 | Ldb1 | 4797 | 0.303 | 0.1945 | Yes |
| 7 | Plg | 5134 | 0.286 | 0.2218 | Yes |
| 8 | Nrp1 | 5447 | 0.274 | 0.2483 | Yes |
| 9 | Ophn1 | 6350 | 0.231 | 0.2475 | Yes |
| 10 | Tle1 | 7197 | 0.205 | 0.2452 | Yes |
| 11 | Pml | 7286 | 0.202 | 0.2699 | Yes |
| 12 | Rtn1 | 10464 | 0.101 | 0.1687 | No |
| 13 | Cdk5r1 | 10919 | 0.086 | 0.1641 | No |
| 14 | Slit1 | 11157 | 0.078 | 0.1662 | No |
| 15 | Dpysl2 | 11269 | 0.073 | 0.1723 | No |
| 16 | Rasa1 | 12534 | 0.032 | 0.1309 | No |
| 17 | Cntfr | 13818 | 0.000 | 0.0843 | No |
| 18 | Ptch1 | 15590 | -0.050 | 0.0269 | No |
| 19 | Vegfa | 15648 | -0.051 | 0.0320 | No |
| 20 | Nf1 | 15677 | -0.052 | 0.0382 | No |
| 21 | Nrp2 | 16486 | -0.075 | 0.0193 | No |
| 22 | Unc5c | 16717 | -0.083 | 0.0224 | No |
| 23 | Tle3 | 17131 | -0.098 | 0.0210 | No |
| 24 | Adgrg1 | 17662 | -0.118 | 0.0181 | No |
| 25 | Vldlr | 19524 | -0.184 | -0.0240 | No |
| 26 | L1cam | 21490 | -0.251 | -0.0606 | No |
| 27 | Hey2 | 21996 | -0.269 | -0.0418 | No |
| 28 | Ets2 | 22010 | -0.270 | -0.0050 | No |
| 29 | Gli1 | 22209 | -0.279 | 0.0263 | No |
| 30 | Crmp1 | 22256 | -0.281 | 0.0634 | No |
| 31 | Nrcam | 22571 | -0.292 | 0.0924 | No |
| 32 | Hey1 | 23117 | -0.314 | 0.1160 | No |
| 33 | Cdk6 | 23589 | -0.338 | 0.1456 | No |