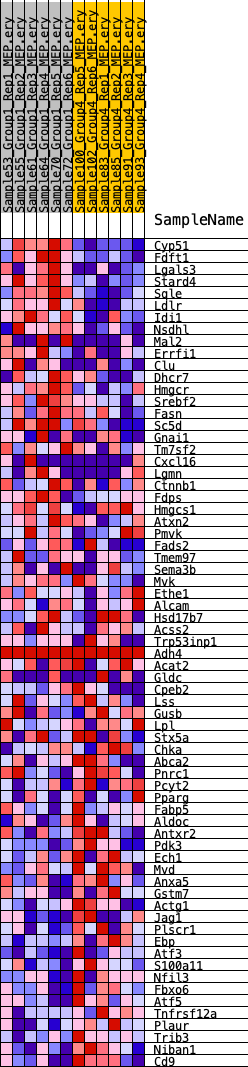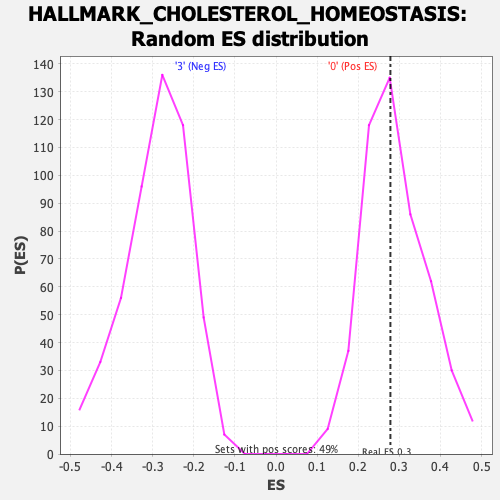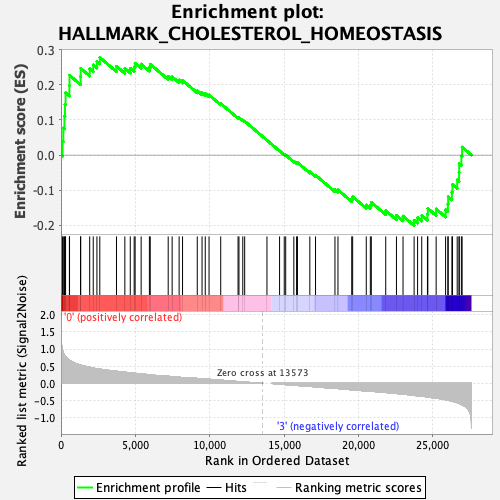
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group4.MEP.ery_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.2782334 |
| Normalized Enrichment Score (NES) | 0.963051 |
| Nominal p-value | 0.51124746 |
| FDR q-value | 0.9969184 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cyp51 | 103 | 0.980 | 0.0395 | Yes |
| 2 | Fdft1 | 154 | 0.900 | 0.0774 | Yes |
| 3 | Lgals3 | 232 | 0.820 | 0.1108 | Yes |
| 4 | Stard4 | 267 | 0.796 | 0.1447 | Yes |
| 5 | Sqle | 307 | 0.779 | 0.1776 | Yes |
| 6 | Ldlr | 565 | 0.674 | 0.1980 | Yes |
| 7 | Idi1 | 573 | 0.672 | 0.2275 | Yes |
| 8 | Nsdhl | 1314 | 0.532 | 0.2240 | Yes |
| 9 | Mal2 | 1325 | 0.530 | 0.2471 | Yes |
| 10 | Errfi1 | 1930 | 0.469 | 0.2458 | Yes |
| 11 | Clu | 2168 | 0.445 | 0.2569 | Yes |
| 12 | Dhcr7 | 2415 | 0.429 | 0.2669 | Yes |
| 13 | Hmgcr | 2609 | 0.416 | 0.2782 | Yes |
| 14 | Srebf2 | 3731 | 0.352 | 0.2530 | No |
| 15 | Fasn | 4293 | 0.325 | 0.2470 | No |
| 16 | Sc5d | 4658 | 0.309 | 0.2474 | No |
| 17 | Gnai1 | 4921 | 0.295 | 0.2509 | No |
| 18 | Tm7sf2 | 4984 | 0.293 | 0.2616 | No |
| 19 | Cxcl16 | 5389 | 0.277 | 0.2592 | No |
| 20 | Lgmn | 5944 | 0.249 | 0.2500 | No |
| 21 | Ctnnb1 | 6001 | 0.247 | 0.2589 | No |
| 22 | Fdps | 7208 | 0.205 | 0.2241 | No |
| 23 | Hmgcs1 | 7474 | 0.195 | 0.2231 | No |
| 24 | Atxn2 | 7943 | 0.176 | 0.2139 | No |
| 25 | Pmvk | 8173 | 0.168 | 0.2130 | No |
| 26 | Fads2 | 9163 | 0.143 | 0.1834 | No |
| 27 | Tmem97 | 9484 | 0.133 | 0.1776 | No |
| 28 | Sema3b | 9700 | 0.126 | 0.1753 | No |
| 29 | Mvk | 9955 | 0.118 | 0.1713 | No |
| 30 | Ethe1 | 10740 | 0.091 | 0.1469 | No |
| 31 | Alcam | 11906 | 0.053 | 0.1069 | No |
| 32 | Hsd17b7 | 11966 | 0.050 | 0.1070 | No |
| 33 | Acss2 | 12223 | 0.042 | 0.0996 | No |
| 34 | Trp53inp1 | 12351 | 0.038 | 0.0966 | No |
| 35 | Adh4 | 13849 | 0.000 | 0.0422 | No |
| 36 | Acat2 | 14698 | -0.018 | 0.0122 | No |
| 37 | Gldc | 15012 | -0.030 | 0.0022 | No |
| 38 | Cpeb2 | 15109 | -0.033 | 0.0002 | No |
| 39 | Lss | 15657 | -0.052 | -0.0174 | No |
| 40 | Gusb | 15841 | -0.057 | -0.0215 | No |
| 41 | Lpl | 15900 | -0.059 | -0.0210 | No |
| 42 | Stx5a | 16732 | -0.084 | -0.0475 | No |
| 43 | Chka | 17116 | -0.098 | -0.0571 | No |
| 44 | Abca2 | 18422 | -0.140 | -0.0983 | No |
| 45 | Pnrc1 | 18624 | -0.147 | -0.0991 | No |
| 46 | Pcyt2 | 19550 | -0.185 | -0.1246 | No |
| 47 | Pparg | 19612 | -0.188 | -0.1185 | No |
| 48 | Fabp5 | 20527 | -0.218 | -0.1420 | No |
| 49 | Aldoc | 20801 | -0.222 | -0.1422 | No |
| 50 | Antxr2 | 20866 | -0.226 | -0.1345 | No |
| 51 | Pdk3 | 21836 | -0.262 | -0.1582 | No |
| 52 | Ech1 | 22557 | -0.292 | -0.1715 | No |
| 53 | Mvd | 22999 | -0.311 | -0.1738 | No |
| 54 | Anxa5 | 23745 | -0.347 | -0.1855 | No |
| 55 | Gstm7 | 23978 | -0.359 | -0.1781 | No |
| 56 | Actg1 | 24257 | -0.371 | -0.1719 | No |
| 57 | Jag1 | 24641 | -0.392 | -0.1685 | No |
| 58 | Plscr1 | 24667 | -0.394 | -0.1520 | No |
| 59 | Ebp | 25226 | -0.426 | -0.1535 | No |
| 60 | Atf3 | 25861 | -0.473 | -0.1556 | No |
| 61 | S100a11 | 26015 | -0.488 | -0.1396 | No |
| 62 | Nfil3 | 26035 | -0.490 | -0.1187 | No |
| 63 | Fbxo6 | 26287 | -0.514 | -0.1052 | No |
| 64 | Atf5 | 26327 | -0.518 | -0.0837 | No |
| 65 | Tnfrsf12a | 26642 | -0.558 | -0.0705 | No |
| 66 | Plaur | 26767 | -0.578 | -0.0495 | No |
| 67 | Trib3 | 26777 | -0.579 | -0.0243 | No |
| 68 | Niban1 | 26923 | -0.609 | -0.0027 | No |
| 69 | Cd9 | 26972 | -0.620 | 0.0229 | No |