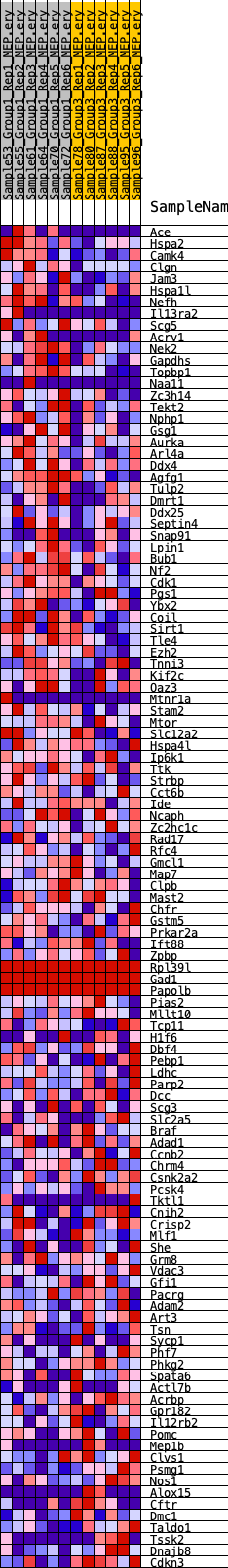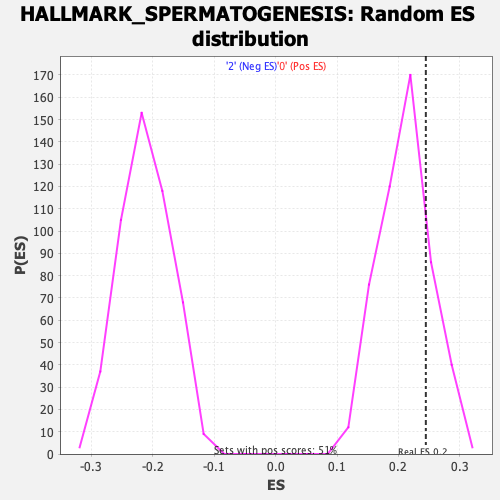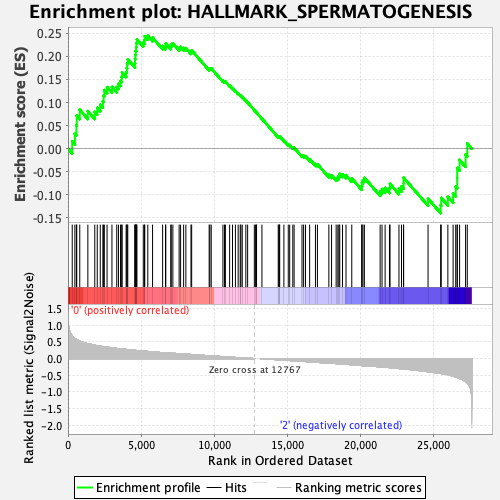
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group3.MEP.ery_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.24452524 |
| Normalized Enrichment Score (NES) | 1.1609508 |
| Nominal p-value | 0.20315582 |
| FDR q-value | 0.4819727 |
| FWER p-Value | 0.965 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ace | 289 | 0.678 | 0.0157 | Yes |
| 2 | Hspa2 | 470 | 0.600 | 0.0323 | Yes |
| 3 | Camk4 | 575 | 0.570 | 0.0505 | Yes |
| 4 | Clgn | 598 | 0.564 | 0.0715 | Yes |
| 5 | Jam3 | 804 | 0.524 | 0.0843 | Yes |
| 6 | Hspa1l | 1357 | 0.445 | 0.0814 | Yes |
| 7 | Nefh | 1830 | 0.400 | 0.0797 | Yes |
| 8 | Il13ra2 | 1999 | 0.386 | 0.0885 | Yes |
| 9 | Scg5 | 2205 | 0.373 | 0.0954 | Yes |
| 10 | Acrv1 | 2384 | 0.362 | 0.1029 | Yes |
| 11 | Nek2 | 2434 | 0.359 | 0.1150 | Yes |
| 12 | Gapdhs | 2482 | 0.356 | 0.1270 | Yes |
| 13 | Topbp1 | 2670 | 0.340 | 0.1333 | Yes |
| 14 | Naa11 | 3004 | 0.324 | 0.1337 | Yes |
| 15 | Zc3h14 | 3332 | 0.305 | 0.1336 | Yes |
| 16 | Tekt2 | 3449 | 0.297 | 0.1409 | Yes |
| 17 | Nphp1 | 3591 | 0.290 | 0.1470 | Yes |
| 18 | Gsg1 | 3671 | 0.287 | 0.1552 | Yes |
| 19 | Aurka | 3698 | 0.285 | 0.1652 | Yes |
| 20 | Arl4a | 3974 | 0.275 | 0.1658 | Yes |
| 21 | Ddx4 | 4022 | 0.273 | 0.1747 | Yes |
| 22 | Agfg1 | 4041 | 0.272 | 0.1845 | Yes |
| 23 | Tulp2 | 4086 | 0.270 | 0.1933 | Yes |
| 24 | Dmrt1 | 4572 | 0.248 | 0.1853 | Yes |
| 25 | Ddx25 | 4584 | 0.248 | 0.1944 | Yes |
| 26 | Septin4 | 4598 | 0.247 | 0.2035 | Yes |
| 27 | Snap91 | 4628 | 0.246 | 0.2119 | Yes |
| 28 | Lpin1 | 4665 | 0.244 | 0.2200 | Yes |
| 29 | Bub1 | 4674 | 0.243 | 0.2291 | Yes |
| 30 | Nf2 | 4712 | 0.241 | 0.2371 | Yes |
| 31 | Cdk1 | 5146 | 0.226 | 0.2301 | Yes |
| 32 | Pgs1 | 5232 | 0.225 | 0.2356 | Yes |
| 33 | Ybx2 | 5245 | 0.224 | 0.2438 | Yes |
| 34 | Coil | 5455 | 0.214 | 0.2445 | Yes |
| 35 | Sirt1 | 5780 | 0.203 | 0.2406 | No |
| 36 | Tle4 | 6468 | 0.177 | 0.2224 | No |
| 37 | Ezh2 | 6666 | 0.171 | 0.2219 | No |
| 38 | Tnni3 | 6684 | 0.171 | 0.2278 | No |
| 39 | Kif2c | 7026 | 0.159 | 0.2216 | No |
| 40 | Oaz3 | 7055 | 0.157 | 0.2266 | No |
| 41 | Mtnr1a | 7168 | 0.155 | 0.2286 | No |
| 42 | Stam2 | 7596 | 0.146 | 0.2187 | No |
| 43 | Mtor | 7697 | 0.143 | 0.2206 | No |
| 44 | Slc12a2 | 7903 | 0.137 | 0.2184 | No |
| 45 | Hspa4l | 8057 | 0.133 | 0.2180 | No |
| 46 | Ip6k1 | 8406 | 0.122 | 0.2100 | No |
| 47 | Ttk | 8453 | 0.120 | 0.2130 | No |
| 48 | Strbp | 9649 | 0.085 | 0.1728 | No |
| 49 | Cct6b | 9697 | 0.083 | 0.1743 | No |
| 50 | Ide | 9790 | 0.081 | 0.1741 | No |
| 51 | Ncaph | 10590 | 0.058 | 0.1473 | No |
| 52 | Zc2hc1c | 10698 | 0.055 | 0.1455 | No |
| 53 | Rad17 | 10758 | 0.053 | 0.1454 | No |
| 54 | Rfc4 | 11049 | 0.045 | 0.1366 | No |
| 55 | Gmcl1 | 11244 | 0.039 | 0.1310 | No |
| 56 | Map7 | 11443 | 0.033 | 0.1251 | No |
| 57 | Clpb | 11631 | 0.029 | 0.1194 | No |
| 58 | Mast2 | 11771 | 0.026 | 0.1154 | No |
| 59 | Chfr | 11819 | 0.025 | 0.1147 | No |
| 60 | Gstm5 | 11910 | 0.023 | 0.1123 | No |
| 61 | Prkar2a | 12156 | 0.017 | 0.1040 | No |
| 62 | Ift88 | 12279 | 0.013 | 0.1000 | No |
| 63 | Zpbp | 12736 | 0.001 | 0.0835 | No |
| 64 | Rpl39l | 12822 | 0.000 | 0.0804 | No |
| 65 | Gad1 | 12875 | 0.000 | 0.0785 | No |
| 66 | Papolb | 12888 | 0.000 | 0.0781 | No |
| 67 | Pias2 | 13256 | -0.003 | 0.0648 | No |
| 68 | Mllt10 | 14370 | -0.035 | 0.0257 | No |
| 69 | Tcp11 | 14418 | -0.036 | 0.0254 | No |
| 70 | H1f6 | 14441 | -0.037 | 0.0260 | No |
| 71 | Dbf4 | 14470 | -0.037 | 0.0264 | No |
| 72 | Pebp1 | 14757 | -0.046 | 0.0178 | No |
| 73 | Ldhc | 15056 | -0.054 | 0.0090 | No |
| 74 | Parp2 | 15154 | -0.057 | 0.0077 | No |
| 75 | Dcc | 15367 | -0.065 | 0.0025 | No |
| 76 | Scg3 | 15455 | -0.066 | 0.0019 | No |
| 77 | Slc2a5 | 15995 | -0.082 | -0.0146 | No |
| 78 | Braf | 16106 | -0.084 | -0.0153 | No |
| 79 | Adad1 | 16235 | -0.088 | -0.0166 | No |
| 80 | Ccnb2 | 16518 | -0.096 | -0.0231 | No |
| 81 | Chrm4 | 16914 | -0.109 | -0.0333 | No |
| 82 | Csnk2a2 | 17048 | -0.113 | -0.0338 | No |
| 83 | Pcsk4 | 17830 | -0.135 | -0.0569 | No |
| 84 | Tktl1 | 18014 | -0.140 | -0.0582 | No |
| 85 | Cnih2 | 18329 | -0.149 | -0.0639 | No |
| 86 | Crisp2 | 18445 | -0.152 | -0.0622 | No |
| 87 | Mlf1 | 18518 | -0.154 | -0.0589 | No |
| 88 | She | 18576 | -0.157 | -0.0549 | No |
| 89 | Grm8 | 18765 | -0.163 | -0.0554 | No |
| 90 | Vdac3 | 19003 | -0.171 | -0.0575 | No |
| 91 | Gfi1 | 19397 | -0.184 | -0.0646 | No |
| 92 | Pacrg | 20075 | -0.208 | -0.0812 | No |
| 93 | Adam2 | 20095 | -0.209 | -0.0738 | No |
| 94 | Art3 | 20162 | -0.212 | -0.0681 | No |
| 95 | Tsn | 20258 | -0.215 | -0.0632 | No |
| 96 | Sycp1 | 21336 | -0.249 | -0.0928 | No |
| 97 | Phf7 | 21465 | -0.255 | -0.0876 | No |
| 98 | Phkg2 | 21676 | -0.261 | -0.0852 | No |
| 99 | Spata6 | 21974 | -0.270 | -0.0855 | No |
| 100 | Actl7b | 22012 | -0.271 | -0.0764 | No |
| 101 | Acrbp | 22622 | -0.298 | -0.0870 | No |
| 102 | Gpr182 | 22796 | -0.301 | -0.0817 | No |
| 103 | Il12rb2 | 22937 | -0.308 | -0.0749 | No |
| 104 | Pomc | 22939 | -0.309 | -0.0630 | No |
| 105 | Mep1b | 24611 | -0.395 | -0.1086 | No |
| 106 | Clvs1 | 25471 | -0.444 | -0.1227 | No |
| 107 | Psmg1 | 25512 | -0.448 | -0.1068 | No |
| 108 | Nos1 | 25965 | -0.485 | -0.1046 | No |
| 109 | Alox15 | 26323 | -0.528 | -0.0971 | No |
| 110 | Cftr | 26502 | -0.555 | -0.0822 | No |
| 111 | Dmc1 | 26603 | -0.570 | -0.0638 | No |
| 112 | Taldo1 | 26605 | -0.570 | -0.0418 | No |
| 113 | Tssk2 | 26767 | -0.601 | -0.0245 | No |
| 114 | Dnajb8 | 27177 | -0.689 | -0.0127 | No |
| 115 | Cdkn3 | 27292 | -0.729 | 0.0113 | No |