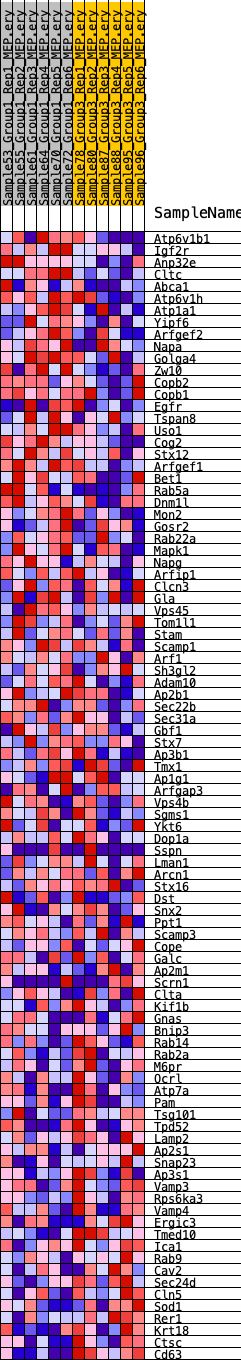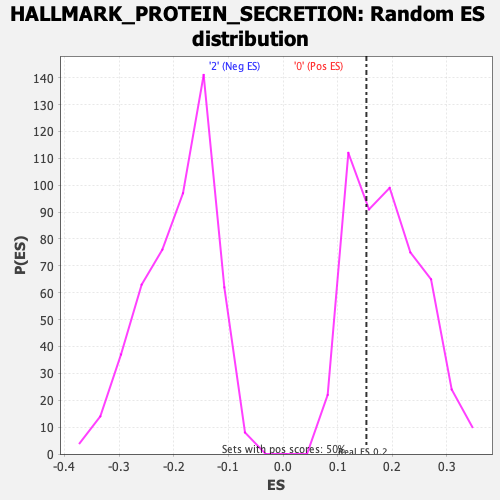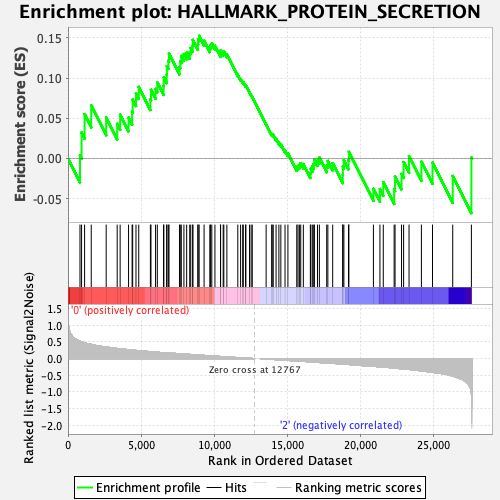
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group3.MEP.ery_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.15259285 |
| Normalized Enrichment Score (NES) | 0.8063163 |
| Nominal p-value | 0.6566265 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp6v1b1 | 819 | 0.521 | 0.0037 | Yes |
| 2 | Igf2r | 927 | 0.506 | 0.0323 | Yes |
| 3 | Anp32e | 1134 | 0.473 | 0.0552 | Yes |
| 4 | Cltc | 1588 | 0.426 | 0.0660 | Yes |
| 5 | Abca1 | 2610 | 0.345 | 0.0510 | Yes |
| 6 | Atp6v1h | 3362 | 0.302 | 0.0431 | Yes |
| 7 | Atp1a1 | 3564 | 0.292 | 0.0546 | Yes |
| 8 | Yipf6 | 4139 | 0.267 | 0.0508 | Yes |
| 9 | Arfgef2 | 4382 | 0.256 | 0.0584 | Yes |
| 10 | Napa | 4421 | 0.254 | 0.0734 | Yes |
| 11 | Golga4 | 4647 | 0.244 | 0.0808 | Yes |
| 12 | Zw10 | 4836 | 0.235 | 0.0891 | Yes |
| 13 | Copb2 | 5631 | 0.208 | 0.0736 | Yes |
| 14 | Copb1 | 5671 | 0.207 | 0.0855 | Yes |
| 15 | Egfr | 5985 | 0.195 | 0.0866 | Yes |
| 16 | Tspan8 | 6105 | 0.190 | 0.0945 | Yes |
| 17 | Uso1 | 6534 | 0.175 | 0.0902 | Yes |
| 18 | Cog2 | 6549 | 0.174 | 0.1009 | Yes |
| 19 | Stx12 | 6746 | 0.168 | 0.1045 | Yes |
| 20 | Arfgef1 | 6759 | 0.167 | 0.1148 | Yes |
| 21 | Bet1 | 6858 | 0.164 | 0.1218 | Yes |
| 22 | Rab5a | 6904 | 0.163 | 0.1306 | Yes |
| 23 | Dnm1l | 7620 | 0.146 | 0.1139 | Yes |
| 24 | Mon2 | 7673 | 0.144 | 0.1213 | Yes |
| 25 | Gosr2 | 7745 | 0.141 | 0.1278 | Yes |
| 26 | Rab22a | 7923 | 0.136 | 0.1301 | Yes |
| 27 | Mapk1 | 8111 | 0.131 | 0.1317 | Yes |
| 28 | Napg | 8321 | 0.125 | 0.1321 | Yes |
| 29 | Arfip1 | 8389 | 0.123 | 0.1376 | Yes |
| 30 | Clcn3 | 8527 | 0.118 | 0.1402 | Yes |
| 31 | Gla | 8532 | 0.118 | 0.1476 | Yes |
| 32 | Vps45 | 8875 | 0.108 | 0.1421 | Yes |
| 33 | Tom1l1 | 8891 | 0.108 | 0.1485 | Yes |
| 34 | Stam | 8965 | 0.105 | 0.1526 | Yes |
| 35 | Scamp1 | 9302 | 0.096 | 0.1465 | No |
| 36 | Arf1 | 9694 | 0.083 | 0.1377 | No |
| 37 | Sh3gl2 | 9746 | 0.082 | 0.1411 | No |
| 38 | Adam10 | 9829 | 0.080 | 0.1432 | No |
| 39 | Ap2b1 | 10044 | 0.074 | 0.1402 | No |
| 40 | Sec22b | 10427 | 0.063 | 0.1303 | No |
| 41 | Sec31a | 10428 | 0.063 | 0.1343 | No |
| 42 | Gbf1 | 10607 | 0.057 | 0.1316 | No |
| 43 | Stx7 | 10652 | 0.056 | 0.1336 | No |
| 44 | Ap3b1 | 10860 | 0.050 | 0.1292 | No |
| 45 | Tmx1 | 11607 | 0.030 | 0.1040 | No |
| 46 | Ap1g1 | 11797 | 0.025 | 0.0988 | No |
| 47 | Arfgap3 | 11943 | 0.022 | 0.0949 | No |
| 48 | Vps4b | 11979 | 0.021 | 0.0950 | No |
| 49 | Sgms1 | 12137 | 0.017 | 0.0904 | No |
| 50 | Ykt6 | 12158 | 0.017 | 0.0908 | No |
| 51 | Dop1a | 12409 | 0.009 | 0.0823 | No |
| 52 | Sspn | 12518 | 0.006 | 0.0787 | No |
| 53 | Lman1 | 12595 | 0.004 | 0.0762 | No |
| 54 | Arcn1 | 13543 | -0.011 | 0.0425 | No |
| 55 | Stx16 | 13922 | -0.022 | 0.0301 | No |
| 56 | Dst | 13956 | -0.023 | 0.0304 | No |
| 57 | Snx2 | 14029 | -0.025 | 0.0294 | No |
| 58 | Ppt1 | 14222 | -0.031 | 0.0244 | No |
| 59 | Scamp3 | 14415 | -0.036 | 0.0197 | No |
| 60 | Cope | 14549 | -0.040 | 0.0174 | No |
| 61 | Galc | 14834 | -0.048 | 0.0101 | No |
| 62 | Ap2m1 | 15032 | -0.054 | 0.0064 | No |
| 63 | Scrn1 | 15636 | -0.072 | -0.0109 | No |
| 64 | Clta | 15721 | -0.074 | -0.0092 | No |
| 65 | Kif1b | 15817 | -0.077 | -0.0077 | No |
| 66 | Gnas | 15908 | -0.080 | -0.0058 | No |
| 67 | Bnip3 | 16086 | -0.084 | -0.0069 | No |
| 68 | Rab14 | 16566 | -0.098 | -0.0180 | No |
| 69 | Rab2a | 16595 | -0.099 | -0.0127 | No |
| 70 | M6pr | 16701 | -0.102 | -0.0100 | No |
| 71 | Ocrl | 16798 | -0.105 | -0.0067 | No |
| 72 | Atp7a | 16841 | -0.106 | -0.0014 | No |
| 73 | Pam | 17053 | -0.113 | -0.0018 | No |
| 74 | Tsg101 | 17182 | -0.117 | 0.0011 | No |
| 75 | Tpd52 | 17684 | -0.131 | -0.0088 | No |
| 76 | Lamp2 | 17762 | -0.133 | -0.0030 | No |
| 77 | Ap2s1 | 18092 | -0.141 | -0.0059 | No |
| 78 | Snap23 | 18779 | -0.163 | -0.0204 | No |
| 79 | Ap3s1 | 18786 | -0.164 | -0.0101 | No |
| 80 | Vamp3 | 18857 | -0.166 | -0.0020 | No |
| 81 | Rps6ka3 | 19173 | -0.176 | -0.0022 | No |
| 82 | Vamp4 | 19200 | -0.177 | 0.0082 | No |
| 83 | Ergic3 | 20876 | -0.231 | -0.0378 | No |
| 84 | Tmed10 | 21322 | -0.248 | -0.0380 | No |
| 85 | Ica1 | 21551 | -0.258 | -0.0298 | No |
| 86 | Rab9 | 22293 | -0.283 | -0.0385 | No |
| 87 | Cav2 | 22355 | -0.286 | -0.0224 | No |
| 88 | Sec24d | 22798 | -0.301 | -0.0192 | No |
| 89 | Cln5 | 22940 | -0.309 | -0.0045 | No |
| 90 | Sod1 | 23311 | -0.326 | 0.0029 | No |
| 91 | Rer1 | 24155 | -0.370 | -0.0040 | No |
| 92 | Krt18 | 24915 | -0.411 | -0.0052 | No |
| 93 | Ctsc | 26298 | -0.525 | -0.0218 | No |
| 94 | Cd63 | 27573 | -1.078 | 0.0011 | No |