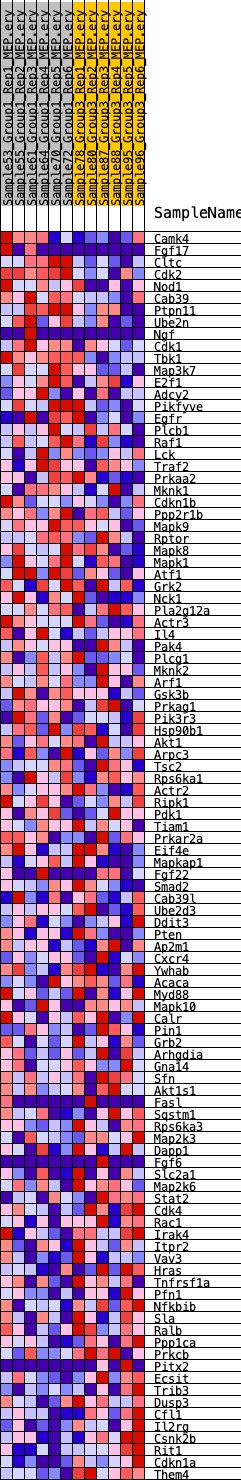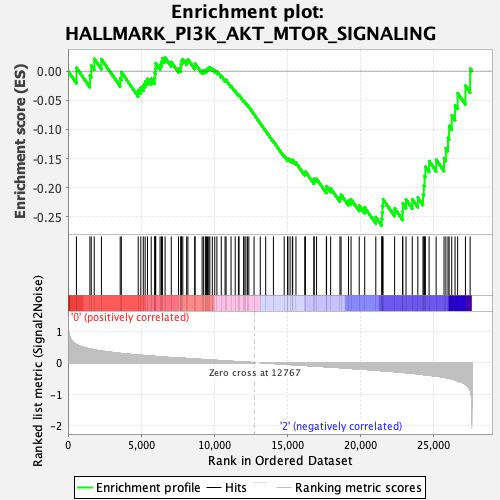
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.ery_Pheno.cls #Group1_versus_Group3.MEP.ery_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.ery_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.2649839 |
| Normalized Enrichment Score (NES) | -1.1896765 |
| Nominal p-value | 0.27290836 |
| FDR q-value | 0.53491026 |
| FWER p-Value | 0.941 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Camk4 | 575 | 0.570 | 0.0059 | No |
| 2 | Fgf17 | 1498 | 0.432 | -0.0073 | No |
| 3 | Cltc | 1588 | 0.426 | 0.0095 | No |
| 4 | Cdk2 | 1792 | 0.404 | 0.0211 | No |
| 5 | Nod1 | 2285 | 0.367 | 0.0205 | No |
| 6 | Cab39 | 3568 | 0.292 | -0.0124 | No |
| 7 | Ptpn11 | 3646 | 0.288 | -0.0017 | No |
| 8 | Ube2n | 4799 | 0.237 | -0.0324 | No |
| 9 | Ngf | 4977 | 0.229 | -0.0280 | No |
| 10 | Cdk1 | 5146 | 0.226 | -0.0235 | No |
| 11 | Tbk1 | 5273 | 0.222 | -0.0176 | No |
| 12 | Map3k7 | 5431 | 0.215 | -0.0132 | No |
| 13 | E2f1 | 5689 | 0.207 | -0.0128 | No |
| 14 | Adcy2 | 5924 | 0.198 | -0.0120 | No |
| 15 | Pikfyve | 5938 | 0.197 | -0.0032 | No |
| 16 | Egfr | 5985 | 0.195 | 0.0043 | No |
| 17 | Plcb1 | 5994 | 0.195 | 0.0132 | No |
| 18 | Raf1 | 6294 | 0.183 | 0.0109 | No |
| 19 | Lck | 6387 | 0.180 | 0.0160 | No |
| 20 | Traf2 | 6455 | 0.178 | 0.0220 | No |
| 21 | Prkaa2 | 6636 | 0.172 | 0.0235 | No |
| 22 | Mknk1 | 7059 | 0.157 | 0.0156 | No |
| 23 | Cdkn1b | 7552 | 0.148 | 0.0046 | No |
| 24 | Ppp2r1b | 7711 | 0.143 | 0.0056 | No |
| 25 | Mapk9 | 7723 | 0.142 | 0.0119 | No |
| 26 | Rptor | 7764 | 0.141 | 0.0170 | No |
| 27 | Mapk8 | 7848 | 0.138 | 0.0205 | No |
| 28 | Mapk1 | 8111 | 0.131 | 0.0172 | No |
| 29 | Atf1 | 8195 | 0.129 | 0.0202 | No |
| 30 | Grk2 | 8660 | 0.114 | 0.0087 | No |
| 31 | Nck1 | 8690 | 0.113 | 0.0130 | No |
| 32 | Pla2g12a | 9167 | 0.100 | 0.0003 | No |
| 33 | Actr3 | 9272 | 0.097 | 0.0011 | No |
| 34 | Il4 | 9392 | 0.093 | 0.0011 | No |
| 35 | Pak4 | 9466 | 0.091 | 0.0027 | No |
| 36 | Plcg1 | 9534 | 0.088 | 0.0045 | No |
| 37 | Mknk2 | 9618 | 0.086 | 0.0055 | No |
| 38 | Arf1 | 9694 | 0.083 | 0.0067 | No |
| 39 | Gsk3b | 9865 | 0.079 | 0.0042 | No |
| 40 | Prkag1 | 10031 | 0.075 | 0.0017 | No |
| 41 | Pik3r3 | 10178 | 0.070 | -0.0003 | No |
| 42 | Hsp90b1 | 10476 | 0.061 | -0.0082 | No |
| 43 | Akt1 | 10736 | 0.054 | -0.0151 | No |
| 44 | Arpc3 | 10806 | 0.052 | -0.0152 | No |
| 45 | Tsc2 | 11145 | 0.042 | -0.0255 | No |
| 46 | Rps6ka1 | 11428 | 0.034 | -0.0341 | No |
| 47 | Actr2 | 11649 | 0.029 | -0.0408 | No |
| 48 | Ripk1 | 11693 | 0.028 | -0.0411 | No |
| 49 | Pdk1 | 12006 | 0.021 | -0.0515 | No |
| 50 | Tiam1 | 12035 | 0.020 | -0.0515 | No |
| 51 | Prkar2a | 12156 | 0.017 | -0.0551 | No |
| 52 | Eif4e | 12264 | 0.013 | -0.0584 | No |
| 53 | Mapkap1 | 12360 | 0.010 | -0.0613 | No |
| 54 | Fgf22 | 12717 | 0.001 | -0.0742 | No |
| 55 | Smad2 | 13142 | -0.000 | -0.0896 | No |
| 56 | Cab39l | 13512 | -0.010 | -0.1026 | No |
| 57 | Ube2d3 | 14041 | -0.025 | -0.1206 | No |
| 58 | Ddit3 | 14777 | -0.046 | -0.1451 | No |
| 59 | Pten | 15009 | -0.053 | -0.1511 | No |
| 60 | Ap2m1 | 15032 | -0.054 | -0.1493 | No |
| 61 | Cxcr4 | 15179 | -0.058 | -0.1519 | No |
| 62 | Ywhab | 15335 | -0.063 | -0.1546 | No |
| 63 | Acaca | 15359 | -0.064 | -0.1524 | No |
| 64 | Myd88 | 15582 | -0.070 | -0.1572 | No |
| 65 | Mapk10 | 16182 | -0.086 | -0.1749 | No |
| 66 | Calr | 16227 | -0.087 | -0.1724 | No |
| 67 | Pin1 | 16803 | -0.105 | -0.1883 | No |
| 68 | Grb2 | 16829 | -0.106 | -0.1843 | No |
| 69 | Arhgdia | 16989 | -0.111 | -0.1848 | No |
| 70 | Gna14 | 17658 | -0.130 | -0.2030 | No |
| 71 | Sfn | 17674 | -0.131 | -0.1974 | No |
| 72 | Akt1s1 | 17953 | -0.139 | -0.2010 | No |
| 73 | Fasl | 18589 | -0.157 | -0.2167 | No |
| 74 | Sqstm1 | 18665 | -0.159 | -0.2119 | No |
| 75 | Rps6ka3 | 19173 | -0.176 | -0.2221 | No |
| 76 | Map2k3 | 19348 | -0.182 | -0.2198 | No |
| 77 | Dapp1 | 19906 | -0.204 | -0.2305 | No |
| 78 | Fgf6 | 20280 | -0.216 | -0.2339 | No |
| 79 | Slc2a1 | 21039 | -0.237 | -0.2503 | No |
| 80 | Map2k6 | 21443 | -0.254 | -0.2530 | Yes |
| 81 | Stat2 | 21471 | -0.255 | -0.2420 | Yes |
| 82 | Cdk4 | 21496 | -0.256 | -0.2309 | Yes |
| 83 | Rac1 | 21535 | -0.258 | -0.2201 | Yes |
| 84 | Irak4 | 22326 | -0.284 | -0.2355 | Yes |
| 85 | Itpr2 | 22875 | -0.304 | -0.2411 | Yes |
| 86 | Vav3 | 22882 | -0.305 | -0.2270 | Yes |
| 87 | Hras | 23105 | -0.315 | -0.2202 | Yes |
| 88 | Tnfrsf1a | 23532 | -0.336 | -0.2199 | Yes |
| 89 | Pfn1 | 23913 | -0.355 | -0.2170 | Yes |
| 90 | Nfkbib | 24265 | -0.376 | -0.2121 | Yes |
| 91 | Sla | 24320 | -0.380 | -0.1962 | Yes |
| 92 | Ralb | 24371 | -0.383 | -0.1800 | Yes |
| 93 | Ppp1ca | 24427 | -0.387 | -0.1638 | Yes |
| 94 | Prkcb | 24682 | -0.398 | -0.1544 | Yes |
| 95 | Pitx2 | 25164 | -0.423 | -0.1520 | Yes |
| 96 | Ecsit | 25703 | -0.464 | -0.1497 | Yes |
| 97 | Trib3 | 25830 | -0.471 | -0.1321 | Yes |
| 98 | Dusp3 | 25974 | -0.486 | -0.1144 | Yes |
| 99 | Cfl1 | 26051 | -0.494 | -0.0939 | Yes |
| 100 | Il2rg | 26230 | -0.515 | -0.0761 | Yes |
| 101 | Csnk2b | 26457 | -0.548 | -0.0586 | Yes |
| 102 | Rit1 | 26634 | -0.575 | -0.0379 | Yes |
| 103 | Cdkn1a | 27165 | -0.686 | -0.0249 | Yes |
| 104 | Them4 | 27487 | -0.867 | 0.0042 | Yes |